Horticulture & Arboriculture - Juniper Publishers
Abstract
Gramineae, such as Zea mays, Oryza sativa, Sorghum bicolor and Miscanthus, has a high cellulose content in the cell wall. Cellulose synthase gene (CesA) plays an important role in the process of plant growth and cell wall morphogenesis. In this paper, the related research results of CesAs in Gramineae was summarized, and the role of CesAs in regulating cellulose synthesis was analyzed.
Keywords: Gramineae; Cellulose; Cellulose synthase gene
Abbreviations: CesA: Cellulose Synthase; CesA: Cellulose Synthase Gene; CSC: Cellulose Synthase Complex; GT2: Glycosyltransferase 2; PCW: Primary Cell Wall; SCW: Secondary Cell Wall
Introduction
Gramineae have more than 500 genus and more than 8,000 species, which are widely distributed in different ecological environments around the world [1]. Gramineae include important grain or sugar crops such as Oryza sativa, Triticum aestivum, Zea mays and Saccharum officinarum, as well as important biomass ingredients crops such as Miscanthus and Panicum virgatum. Gramineae usually have a high cellulose content [2], and the stability and great ductility of cellulose make it an ideal raw material for manufacturing many fiber products [3,4].
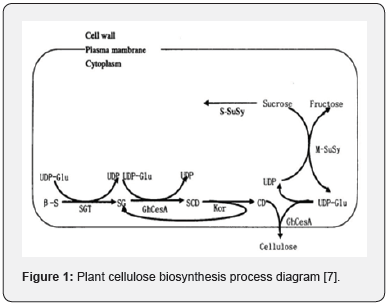
Biosynthesis of Cellulose
Cellulose is one of the important components of the higher plants mature cell walls. As a structural polysaccharide, it accounts for 30-40% of the total cell wall polysaccharides [5]. The basic unit of cellulose is pyranoid D-glucose, which is connected by β-1,4 glucoside bonds and usually exists in the form of microfibrils, and its structure has a specific hydrogen bond structure between molecules [6]. Studies have shown that the pathway of cellulose biosynthesis is mainly divided into three steps (Figure 1), and CesA plays an important role in the program [7]. CesA is a membrane-bound transmembrane protein a member of the glycosyltransferase 2 (GT2) family that catalyzes the formation of β bonds between glycosylates. CesA is a hexagonal lotus shaped enzyme with a diameter of 25-30nm, it can be assembled into a Cellulose Synthase Complex (CSC) with a sixfold symmetrical rosette structure on the cell membrane [8]. The amount of CSC in plants is positively correlated with the final synthesis amount of cellulose. The number of rosette structures of the Arabidopsis mutant rsw1 is significantly less than that of the wild type, which caused β-1,4-glucan to accumulate in an amorphous form and the cellulose content is reduced [9].
Cellulose Synthase Gene (CesA)
The length of CesAs sequence is between 3.5~5.5 kb, containing approximately 9~13 introns and encoding 985~1,088 amino acid [10]. In 1982, Benziman [11]. cloned CesA in vitro bacteria for the first time [11]. In 1996, Peal et al. cloned the β-1,4- glucosidyltransferase gene encoding the catalytic CesA subunit from cotton for the first time by random sequencing and sequence analysis from a cDNA library [12]. Since the discovery of CesA in cotton, CesAs had been successively cloned in Arabidopsis thaliana [13], Oryza sativa [14], Zea mays [15], Populus trichocarpa [16], Boehmeria nivea [17] and Phyllostachys edulis [18]. In the studies of Gramineous CesAs, it was found that OsCesA4, 7,9 of rice were all involved in the formation of SCW, forming a complex regulatory network of SCW cellulose biosynthesis. These three CesA genes can be expressed cooperatively in rice seedling stage, young stem, immature panicle and root development, but relatively little expression in mature leaves [19]. The missense mutation of OsCesA7 caused the decrease of cellulose level and cell wall damage in S1-24 mutant rice; at the heading stage, compared with wild-type, S1-24 mutant rice has a lower mechanical strength and a relatively slower growth rate [20]; however, the expression of OsCesA7 could be directly up-regulated by regulating the MYB transcription factor OsMYB58/63 of rice [21]. When OsCesA9 has a missense mutation, it will cause plant dwarfing and extremely low fertility [22]. In Panicum virgatum L., PvCesA4 and PvCesA6 genes have different expression levels in different parts. After the overexpression and/or knockout of PvCesA4 and PvCesA6, the cellulose content of the transgenic plants decreased, while the xylan content increased. The increase of xylan content would lead to the decrease of crystallinity of cellulose, which would affect the synthesis of cellulose Therefore, the expression of CesAs had changed cell wall composition and cellulose crystallinity [23]. The CesAs of Miscanthus × giganteus has been reported. MgCesA10, MgCesA 11 and MgCesA 12 may participate in the formation of SCW and form an equal proportion of CSC. MgCesA5 and MgCesA6 are constitutively expressed genes that cooperate with MgCesA2, 3, 4, 7 and 8 to regulate the formation of PCW. However, except for MgCesA5, the expression of other CesA genes in leaves was reduced due to senescence. The expression of genes involved in the formation of Miscanthus × giganteus PCW varies depending on the location [24].
Different Regulation Levels of Gramineae CesA
Cellulose synthesis can be regulated at the transcriptional level by CesAs. The biggest difference between CesAs is the presence and location of introns in the coding sequence [13]. For example, wheat CesA1, 2 and 6 have 13 introns, while CesA4, 7 and 8 have 7, 12 and 9 introns respectively; CesA1, 2 and 6 participate in the formation of PCW, while CesA4, 7 and 8 participate in the formation of SCW [25]. This indicated that the number of introns of CesAs in the formation of wheat PCW was higher than that of CesAs in the formation of SCW. Previous studies have shown that genes containing introns have higher transcription levels [26]. This also indicates that transcription levels of CesAs participate in wheat PCW formation are higher than those of CesAs participate in SCW formation. In addition to the regulation of cellulose synthesis at the transcriptional level of CesAs, the post-transcriptional level of CesAs also affects the synthesis of cellulose. Daniel et al. found that the small RNA produced by HvCesA6 can selectively attenuate the expression of CesA gene, therefore, the expression of genes that affect cell wall formation can greatly influence the content of barley cellulose [27].
Conclusion
Cellulose is the most important component of plant cell walls, and cellulose synthase plays a key role in cellulose biosynthesis. In the studies of Gramineae CesAs, it was found that CesA gene family was involved in cell wall morphogenesis, forming a complex regulatory network of cellulose biosynthesis. Transcriptional or post-transcriptional regulation of CesA genes can change plant cell wall composition, change cellulose content and cellulose crystallinity, so as to provide a strong theoretical basis for the high value utilization of cellulosic feedstock crops.To Know more about Horticulture & Arboriculture
Click here: https://juniperpublishers.com/index.php




No comments:
Post a Comment