Cell Science & Molecular Biology - Juniper Publishers
Abstract
The development of pathophysiologies including fibrosis and cancers involve the establishment of a tissue microenvironment characterized by hypoxia and low serum conditions. NHE1 directly influences cell proliferation, extracellular matrix composition, and cell migration in multiple disease states. Calcineurin homologous protein isoforms 1 and 2 (CHP1 & 2) are calcium-binding proteins whose function is not well understood. CHP1 is crucial for NHE1 membrane processing and basal transport activity. Additionally, CHP1 is involved in the regulation of gene expression. In contrast, CHP2 expression is limited to a small range of cells types its primary function is ascribed to the regulation of NHE1. Preferential binding of CHP2 to NHE1 under the tumor microenvironment supports cell survival by first activating the exchanger through the CHP2-NHE1 interaction and by freeing CHP1 to the nucleus where it also supports cell survival. The affinity of CHP2 is three-fold greater than CHP1, but endogenous CHP concentrations indicates that competition alone cannot explain the role of CHP2. Modelling of CHP binding identified two distinct approaches to establishing the CHP-NHE1 interaction; CHP1 binds in a two state-mechanism and CHP2 bound NHE1 in a one state-mechanism. Additional analysis of predicted and published phosphorylation sites of the CHPs suggest that phosphorylation can support a CHP specific interaction with NHE1. Together these data and models highlight a means to identify possible therapies that target CHP2 but not CHP1 to defeat cell survival in several disease states.
Keywords: Sodium Hydrogen Exchanger Isoform 1, NHE1, Calcineurin B homologous protein, CHP1, CHP2, Tumor metastasis, Protein-protein interaction, Tumor microenvironment, TME, targeted therapy, Intracellular pH
Overview
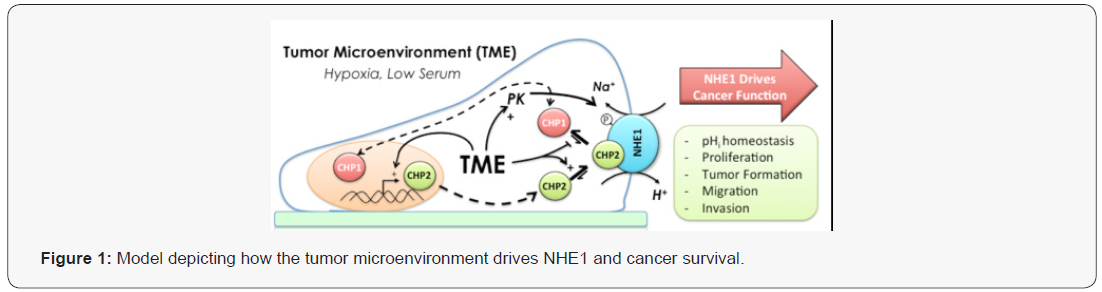
The Na+-H+ exchanger isoform 1 (NHE1) plays a critical role in directing cell functions including proliferation, motility and intracellular pH homeostasis. Poorly understood is how two closely related regulatory proteins, calcineurin B homologous protein 1 and 2 (CHP1 and CHP2), coordinate to control NHE1 related cellular events. Under normal conditions, CHP1 is bound to NHE1 maintaining basal activity. When CHP2 is expressed, it also binds NHE1 at overlapping residues with CHP1 and CHP2 regulates and enhances NHE1 transport. The physiological relevance and regulation of CHP2 has not been elucidated. As visualized in Figure 1, we hypothesize that in the tumour microenvironment, CHP2 displaces CHP1 from NHE1, altering aNHE1 function and freeing CHP1 to locate to the nucleus where it can drive expression of genes supporting cell survival.
NHE1 Regulation and Function
NHE1 belongs to the 13 member SLC9 gene family. Five isoforms, NHE1-5, are located on the plasma membrane and NHE1 is nearly universal in human tissues [1,2] NHE1 has two functions: (i) homeostasis of intra- and extracellular pH through Na+-H+ exchange and (ii) as a scaffold for a range of proteins through protein interactions at an extended cytosolic carboxyl tail. NHE1 regulation is complex and involves several phosphorylation sites, lipid binding domains and protein-protein interactions. How these regulators interact and whether certain forms have priority under different cellular conditions is unknown. Activation of NHE1 is one of the initial steps in the development of the transformed phenotype of cancer cells [3]. This activation leads to an alkalization of intracellular pH (pHi), which is a permissive requirement for cell growth. During cell migration, NHE1 localizes to the leading edge of cells where a combination of an elevated pHi and the binding of a range of proteins to NHE1 stimulates the formation of actin filaments that drive the forward movement of these migrating cells [4-8]. Abnormal regulation and function of NHE1 has been identified as playing a major role of a range of pathophysiological conditions. NHE1 directly influences cell proliferation and migration in multiple disease states. In pulmonary hypertension, pulmonary arterial smooth muscle cells increase proliferation and migration due to the activation of NHE1 in response to chronic hypoxia [9]. In dilated cardiomyopathy, elevated NHE1 activity induces cardiac hypertrophy in vivo driving disease progression [10]. In cancer, activation of NHE1 is an initial step in the development of the transformed phenotype of cancer cells [1-4]. Aberrant activation of NHE1 can also lead to changes in Na+ homeostasis which enhances disease progression. In both perinatal brain injury [11] and diabetic cardiomyopathy [12] anomalous activation is NHE1 drives an increase cytosolic Na+ level which alters Ca++ homeostasis leading to impaired energy metabolism and decreased cell survival. Each of the conditions listed share both the activation of NHE1 as a driving force in the initiation of a disease state and the fact that an underlying mechanism for that NHE1 activation has not been fully elucidated.
Role Of CHP 1 and CHP2
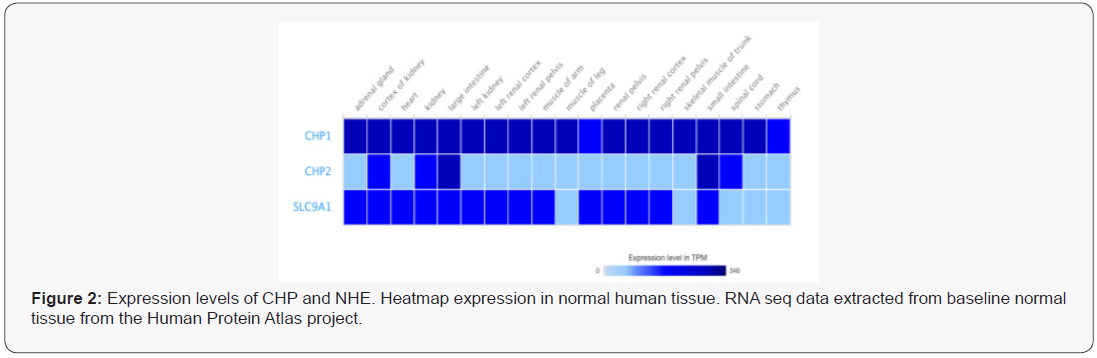
Both CHP isoforms bind and regulate NHE1 transport activity. CHP1 & 2 are calcium-binding proteins whose function is not well understood [13]. CHP1 is crucial for NHE1 membrane processing and basal transport activity. Additionally, CHP1 is involved in the regulation of gene expression [13-15]. No published data relates the distinct nuclear and membrane functions of CHP1 or the mechanisms by which these functions are regulated. In contrast, CHP2 expression is limited to a range of cells types including intestinal and skin cells (Figure 1) and its primary function is ascribed to the regulation of pHi via NHE1 [16]. In non-diseased tissue, CHP2 is expressed at low but detectable levels in most cell types including lung, muscle and other tissues ( Figure 2; RNAseq baseline Human Protein Atlas project: TMP 0.1-0.99), moderately found in kidney, adrenal, prostate, bladder and vagina (TMP 1-9.9) and highly expressed in intestinal track cells, some epidermal cells and spinal cord (TMP => 10.0). We have identified CHP2 expression in several cultured cell lines including CCL39 fibroblasts as well as other transformed lung epithelial and fibroblastic cell lines but not non-diseased cultured cells [17].
CHP1 has several identified functions:
A. at the membrane through binding to NHE1
B. in the cytoplasm engaging in the regulation of microtubule formation and vesicle trafficking
C. in the nucleus where it both positively and negatively regulates gene transcription and nuclear function.
Expression of CHP1 can inhibit, nuclear factor of activated T cells (NFAT) and its associated transcriptional activity by inhibition of calcineurin [18]. Both calcineurin and NFAT are expressed at 2-4 fold higher levels in non-small cell lung cancer (NSCLC) than in non-diseased lung tissue [19]. Hypoxic conditions enhance lung fibroblast proliferation and migration in lung fibrosis while increasing NFAT expression indicating a key role for CHP1 [20]. Both serum deprivation and hypoxia can lead to CHP1 translocation to the nucleus [21]. Nuclear CHP1 has been identified to stabilize HIF1a in hypoxic HepG2 cells inhibiting hypoxia-induced apoptosis [22]. Likewise, the deathassociated protein related apoptosis inducing kinase (DRAK2) binds to and is inhibited by CHP1 and may be involved in both nuclear translocation of CHP1 and inhibiting apoptosis [23]. CHP2 may have an important but unidentified role in several disorders. CHP2 expression was first identified in hepatoma, colon carcinoma, cervical carcinomas, and leukemia cells but not in healthy, non-diseased tissues [24]. Most recently, CHP2 expression was detected as a putative tumour marker in patients with acute leukemia, where it was not detected in paired, nondiseased tissue [25]. While most of the work investigating CHP2 has focused on its regulation of NHE1 in cancer, overexpression of CHP2 activates both calcineurin and NFAT, contrary to the effects of CHP1 [26]. Increased CHP2 expression affords cellular maintenance of an elevated pHi and enhanced cell survival in a low serum environment, as well as an increase in cell proliferation [17]. In addition, in nude mice knockdown of either NHE1 or CHP2 abrogates tumour formation [17]. Overexpression of CHP2 increased cell proliferation, cell adhesion, and cell invasion relative to cells lacking CHP2 expression [27]. Inversely, CHP2 expression knockdown in HepG2 cells inhibited cell proliferation, even in the presence of CHP1 [28]. These data suggest that CHP2 regulation of NHE1 allows a higher basal pHi which supports accelerated rate of proliferation [24].
NHE1-CHP1/2 Interactions As A Therapeutic Target
Current clinical trials using NHE1 inhibitors as therapeutic modalities for several disease have struggled for success, in part, because of the nature of small molecule inhibitors targeting a ubiquitously expressed protein resulting in negative impacts on non-diseased tissue [29,30]. Yet the optimism for an NHE1 related therapy remains high for several disorders as positive results have been observed [31,32]. Data suggests that both CHP1 and CHP2 play a critical role in cell survival, proliferation and motility in microenvironments found in both tumor and fibrotic tissues. Blocking the binding of both CHP isoforms would limit the gain of function that occurs with CHP2 expression but would concomitantly result in loss of CHP1 sequestering at the membrane where it could support diseased cell function in the nucleus. Thus a precision therapy targeting CHP2-NHE1 interactions while sparing CHP1-NHE1 interactions might be a promising approach. Unfortunately, there is currently not enough evidence to justify the development of such a therapy. We next provide evidence for such a CHP-specific approach. While both isoforms of CHP have a 75% amino acid homology with nearly identical binding sites on NHE1[31] there is a fundamental difference in the impact of each CHP-NHE1 interaction on transporter function. CHP1 does not activate NHE1 but its binding is required for agonist activation, while CHP2 increases both the Vmax and the H+ affinity of NHE1, thus serving to activate transport above and beyond CHP1. A single report indicates that CHP2 binds NHE1 with a five-fold greater affinity than CHP1 resulting in significantly increased NHE1 transport activity [33]. In cells stably transfected with CHP2, the resting pHi was 0.2 to 0.3 pH units higher than control cells [34]. No mechanism for the regulation of the CHP-NHE1 interactions has been reported.
CHP-NHE1 Binding
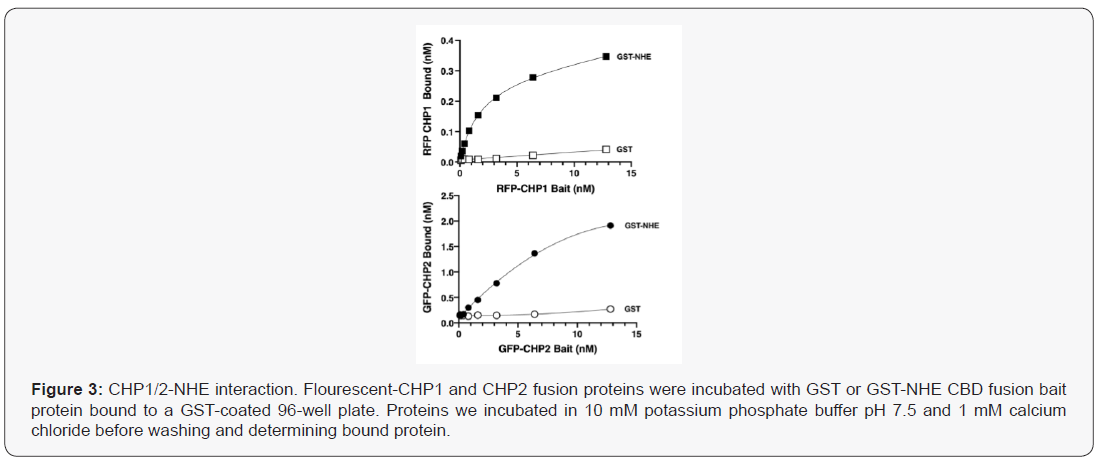
Using a semiquantitative competitive pulldown approach, [24] Pang et.al. demonstrated that CHP2 bound NHE1 with nearly a five times higher binding affinity than CHP1 [33]. These studies indicated that the two CHP isoforms could competitively bind to NHE1 and that concentration might be the driver for binding to NHE1. Recently, the impact of calcium on CHP-NHE1 binding was analysed using isothermal titration calorimetry (ITC; 34). In the absence of calcium, CHP1 binds to NHE1 with a 2.7 times greater affinity than CHP2. Yet, in the presence of calcium, CHP2 but not CHP1 affinity for NHE1 increases by nearly seven-fold, resulting with both isoforms bound to NHE1 with nearly the same affinity (KD = 7.7 nM for CHP1 and 3.2 nM for CHP2-NHE1).To qualitatively examine interactions between NHE1 and CHP, we bound GST or the GST- CHP binding domain of human NHE1 (aa 503-545) to a glutathione coated 96 well plate and determined binding of either RFP-CHP1 or GFP-CHP2 in the presence of 1 mM calcium chloride (Figure 3). Like the ITC generated data, the affinities were in the low molar range (CHP1-NHE1: 2.65 nM, CHP2-NHE1: 9.65 nM) with CHP2 binding NHE with 3.65 fold higher affinity than CHP1. Interestingly, while the KD for each binding interaction was modestly different, the maximal bound CHP significantly differed for each CHP isoform. The potential for an affinity tag to fold over and bind or block the NHE1 binding groove of CHP was suggested after examination of the structure of CHP1 and could be a potential explanation for the differences in bound between CHP1 and CHP2 [17,34]. Using recombinant His tagged purified CHP1/2 and the CHP binding domain of NHE1 after removal of the affinity tag by TEV proteolysis, we determined the melting point for each protein alone or in combination using circular dichroism spectroscopy during a thermal melt. The melting point shift for CHP1-NHE was 3.3 oC greater than for the CHP2-NHE complex (Tm CHP1-NHE1; 58.7 +/- 1.7oC and CHP2-NHE1: 55.4 +/- 1.2oC). The change in free energy (DG at 37oC) of binding for the two complexes was then calculated as -1.16 kJ/mol for CHP1-NHE and -3.91kJ/mol for CHP2-NHE complex. The 2.74kJ/mol difference in predicted binding energy at physiological temperature translates to an increase in affinity of CHP2 over CHP1 by a factor of 3.02. The increased affinity predicted here and identified by pull down assays [33] despite their high level of homology might suggest different binding mechanism for the association and protein recognition.
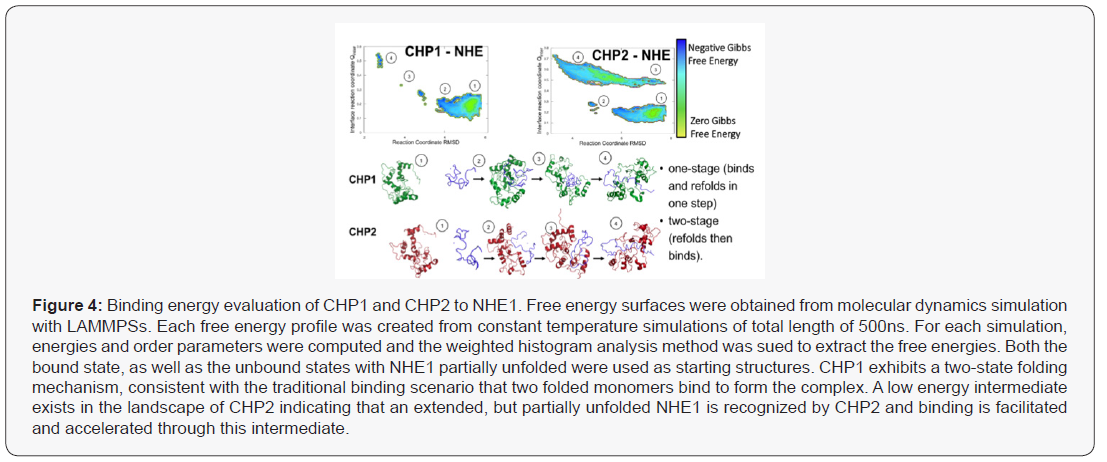
The thermodynamic predictions of binding were analysed using a Weighted Histogram Analysis Method (WHAM) and visualization software (LAMMPS) simulations to calculate binding energies to calculate predicted a binding mechanism (Figure 4). Protein binding landscapes obtained with molecular dynamics simulations show that both CHP1 and CHP2 binding is driven by the overall structure and binding site of the proteins. However, while the landscape of CHP1 exhibits a two-state binding of CHP1 to NHE1, CHP2, even though the monomer is stable on its own, shows binding through a partially unfolded, extended intermediate (Figure 5). Higher binding affinity and specificity through a partially disordered protein, although a bit counter-intuitive at first, has been documented in the literature. The intermediate in the CHP2 energy landscape suggest that the binding interface can be formed early during binding. Overall, the simulation study supports that the binding mechanism is different between CHP1 and CHP2 and CHP2 is accelerated due to the faster recognition of NHE1. Both of these modelled mechanisms are supported by the observed differences in binding between with the two florescent tagged CHP proteins (Figure 3). In this case, the flexible linker between the N-terminus of CHP and GFP/RFP could, as suggested, fold over and sterically block a folded binding site of CHP for NHE1. As modeled, CHP1, binding in a one-state mechanism would already be folded and thus binding events would be blocked but the affinity unchanged, a mechanism similar to a competitive inhibitor reducing Vmax but leaving Km unchanged. Our observation that CHP2 bound with the same affinity as CHP1 to NHE1, but with a higher binding maximum specific binding (Bmax), of 3.55nM for CHP2 and only 0.41 nM for CHP1 would support that the GFP portion of the fusion protein was distal to the unfolded CHP2 peptide which could then bind and fold in the one-state mechanism with less interference than a protein using a one-state binding mechanism.
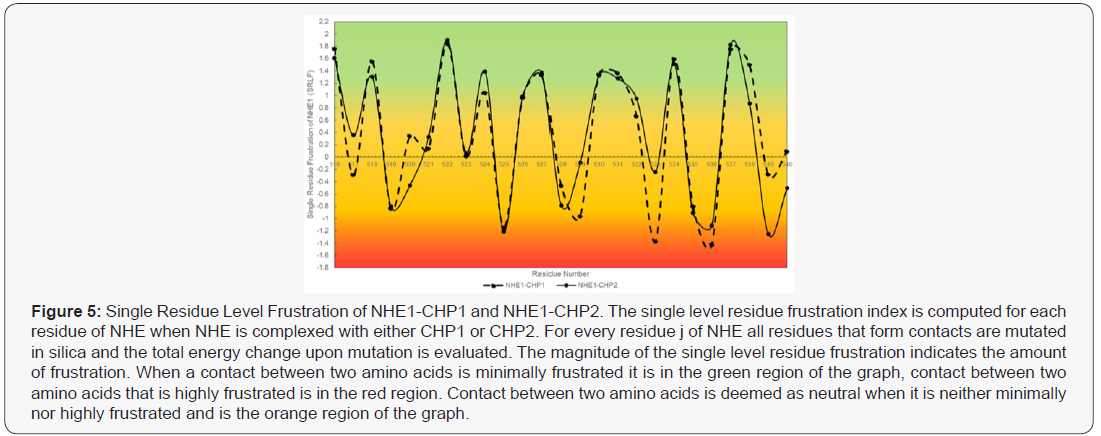
CHP-NHE1 Interaction- Target For Drug Therapies
The structures of CHP1-NHE1 and CHP2-NHE1 have been solved [35,36]. CHP1 and CHP2 form a receiving groove for the NHE1 helix with the hydrophobic side chains of NHE1 aligned to one face making fourteen contact points with CHP. Several of the NHE1 residues are critical for binding CHP and mutation of these amino acids leave the transporter unable to bind either CHP isoform. While CHP isoform specific interactions with NHE1 were not identified, the mutation of NHE1 residues: Phe526, Leu527, Leu530, Leu531, Ile534, or Ile537 were all key for CHP binding and transport function [36,37]. Because CHP1 and CHP2 possess a divergent amino acid change at several of these NHE1 contact sites, it is likely that select NHE1 residues would be potential targets for defining CHP2-NHE1 interactions. Energy frustration computations using the Energy Landscape Theory were performed to quantitate the energy fit of each NHE1 residue making contact with CHP1 or CHP2 (Figure 5) [39,40]. Each NHE residue making contact with CHP1 and CHP2 was analyzed for the degree of local frustration using the freely available resource, EMBnet bioinformatic [41]. This tool scores the energies (local frustrations) of all possible amino acids vs the native amino acids involved in CHP-NHE binding. Additional evidence showing the unique binding of CHP for NHE1 is shown using an overlay of the NHE1 CBD domain bound to CHP1 and CHP2 with only the helix of NHE1 shown (masking CHP structure) (Figure 7) The shift between the two bound conformations of the NHE CBD indicates a subtle yet real difference in the structure of NHE1 as it binds each CHP (Figure 6) . It is possible that subtle contacts which render a specific interaction remain to be elucidated. Such analysis provides insight of biologically important regions of NHE1-CHP binding and identifies 11 sites of that may confer both the differences in binding affinities and highlight possible specific binding sites unique to CHP1 and CHP2.
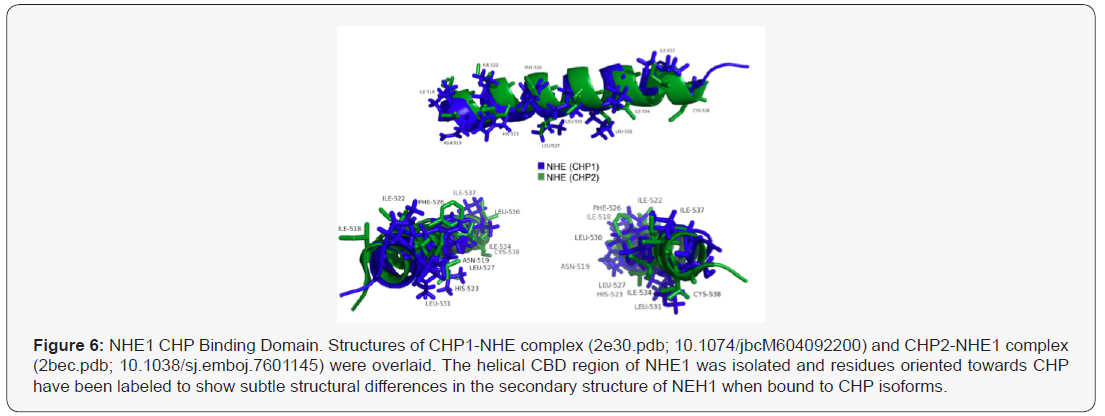
Predictive modelling identifies that while both CHPs bind to nearly the same region of NHE1, there exists potential unique differences of binding of both CHP isoforms. Differences that could be exploited to design a CHP-specific inhibitor. However an argument remains that protein expression and competition alone could drive CHP1-CHP2 binding to NHE1. As seen in our and other published data, the affinity of CHP1 and CHP2 for NHE1 are similar showing a 3-5-fold higher affinity (KD) for CHP2-NHE1. This difference is also demonstrated by the binding models identified here. However, the difference in CHP affinity for NHE1 cannot explain a mechanism for the two CHP isoform binding. Several diseased tissues express higher levels of CHP2 [17,33]. We have shown that CHP2 expression increases in low serum [17] and other tumour microenvironment conditions (data not shown). As CHP2 expression increases due to the changes in the cellular microenvironment, CHP2 is thought to displace CHP1 bound to NHE1, increasing exchanger activity and altering cellular function. Thus at face value, it would seem that increases in protein expression and a slightly higher affinity for CHP2 could explain the phenomena. However, the estimated levels of protein in the highest CHP2 expressing cells favours CHP1 by 20-100 fold in cultured cells and in tissues ranged from 60 to 1000 fold in favor of CHP1 expression (Protein Atlas, GTX, Proteomics DB ; 17, 43). Therefore a model defined by competitive binding is not likely. More attractive is a combination of unique binding contacts and post translational modifications that could serve to regulate binding affinities.
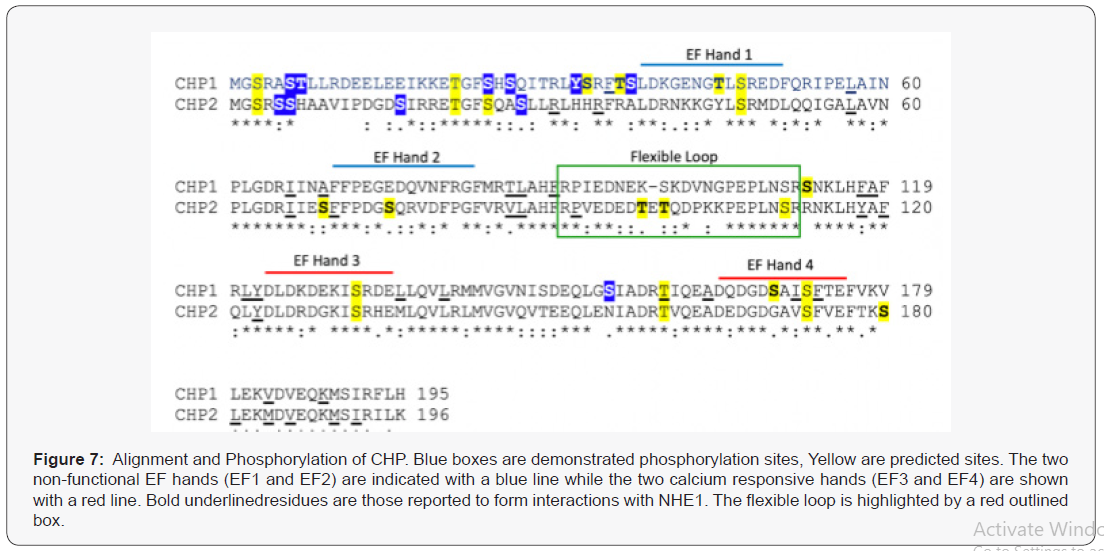
Interestingly, while NHE1 is phosphorylated at 22 putative and known phosphorylation sites [42], the CHP binding domain of NHE1 is not phosphorylated. The nearest modified site to the CBD domain is 50 residues distal to the carboxy-terminus. Both CHP isoforms are highly phosphorylated. Analysis of known phosphorylation sites identified in mass spectroscopy databases shows one site in common for both CHP1 and CHP2 while there are six unique phospho-sites of CHP1 and three unique sites identified for CHP2 [17]. Peptide sequence prediction analysis of NetPhos [43] highlights an additional six unique putative phosphorylation sites for both CHP1 and CHP2 (score, >0.5). There are seven additional sites predicted to be in common for both isoforms (Figure 7). CHP forms a hydrophobic receiving groove for the helical NHE CBD. Interestingly, the C terminal lobe of CHP which is key for binding to NHE1 is devoid of potential phosphorylation sites. The bulk of identified phosphorylation sites are on the N-terminus of the protein which forms the second lobe of CHP but is not a significant component of the NHE1 receiving pocket. Both CHP isoforms have similar C- terminal lobe domains with a unique linker domain at aa 81-101 (Figure 7). The structure of the CBD domain loops out between the N and C lobes when bound to NHE1, suggesting this is an important determination binding factor. Several putative CHP2 specific phosphor-sites are identified in the flexible loop that could be a site of specific regulation. The mechanisms for binding observed in the computer simulations might explain why the N-terminus is nevertheless very important. Despite the stark topological similarity of CHP1 and CHP2 as well as the high sequence homology, binding simulations clearly predict a different mechanism. The different recognition mechanisms suggest an important biological difference between CHP1 and CHP2 on a molecular level, and must be encoded in the proteinprotein interfaces. While the binding free energy landscape of CHP1 follows a more traditional lock-and-key two-state folding mechanism, CHP2 binding exhibits a clear intermediate in the binding pathway. These intermediates can often increase binding affinity by forming recognition-specific contacts. The intermediate found in the CHP2-NHE1 recognition has many of the proteinprotein interface contacts formed fairly early in the binding and recognition process (Figure 3). Interestingly, the intermediate consistently forms strong contacts to multiple residues found in the N-terminus that were predicted to be phosphorylation sites, namely series S3 and S37 (Figure 7) as well as threonine T7 (Figure 7). These residues play an important role in the binding but also in the regulation. A more detailed analysis of binding and regulation specific residues should shed further light on the binding affinity, and could provide a model system for designing therapeutic drugs.
Conclusion
The two proteins, CHP1 and CHP2 each bind to nearly the same site of NHE1 with subtly unique contacts. Endogenous levels of CHP1 are much higher than CHP2 even after expression is induced. While CHP2 has a 3-5 fold higher affinity for NHE1 than CHP2, the concentration of each CHP cannot explain how in particular disease states, CHP2 can displace CHP1 and bind to NHE1 supporting cell behaviour. While critical sites of contact for NHE and CHP have been evaluated, these experiments were all conducted investigating one CHP isoform or the other, never comparing the impact on both isoforms under the same conditions. It is unknown if these mutations would effect one or both CHP proteins. Thus a mechanism for CHP specific binding to NHE1 remains elusive. Because CHP1 has a second role in the nucleus supporting hypoxia and other environmental conditions, it could be possible that CHP2 not only activates NHE1 during disease progression, but CHP2 displacement of CHP1 from the membrane allows CHP1 translocation to the nucleus coordinating cell survival in hypoxic and other disease states. Thus a simple inhibitor non-specifically inhibiting CHP-NHE1 interactions could have the opposite effect by driving CHP1 to the nucleus. Thus what is needed is to further our understanding of how CHP1 and CHP2 each bind and compete for NHE1 binding. This work has identified and summarized possible mechanisms for the development of a new generation of inhibitors of NHE1 function in human disease. Thus, the generation of a functional precision therapy could come by identifying an entity that could block the unique binding interactions between CHP2 and NHE1 or by targeting the distinctive CHP2 phosphorylation sites.
To Know more about Cell Science & Molecular Biology
Click here: https://juniperpublishers.com/index.php




No comments:
Post a Comment