Advances in Biotechnology & Microbiology- Juniper Publishers
Abstract
Proteomics is one of the complex and significant methodologies to know the genetic functions of proteins. The study of plant proteins having antilithiatic activity is an unexplored area and can be hit through proteomics. The present study was framed for in-depth identification and characterization of antilithiatic proteins from horse gram (Macrotyloma uniflorum). Total protein of Macrotyloma uniflorum seeds from six different location of Himachal Pradesh, India was isolated in protease inhibitor buffer. The identification of cultivar seed proteins was done by Matrix-assisted laser desorption/ionization-time of flight Mass spectrometry (MALDI-TOF MS). The results were framed by database search with the MASCOT server. The protein sample of Rampur location contained maximum number of peptides followed by Sundernagar, Palampur and Kullu. Seeds of Chamba location showed the presence of very few peptides. The quantification of Macrotyloma uniflorum seed proteins was done with Bradford method and further protein with high oxalate inhibition activity (15µg) was loaded on 12% Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) gel. In the proteomic study of loaded protein, a total of three major low molecular weight protein bands were observed (~27KDa, ~23.1 and ~17KDa respectively). Seed protein SDS-PAGE may be used to investigate genetic diversity and relatedness in cultivars at a low cost. Furthermore, the proteomic analysis of the seed protein of M. uniflorum aids in the creation of a protein-based marker that can be used to identify cultivars in the future.
Keywords: Proteomics; Macrotyloma uniflorum; MALDI-TOF MS; MASCOT server; Antilithiatic proteins
Abbreviations: MALDI-TOF MS: Matrix-Assisted Laser Desorption/Ionization-Time of Flight-MASS Spectrometry; SDS-PAGE: Sodium Dodecyl Sulphate Polyacrylamide Gel Electrophoresis; TFA: Trifluoroacetic Acid; HCCA: Hydroxycinnamic Acid; CDPK: Calcium-dependent protein kinase 29-like
Introduction
Our urinary system is susceptible to a variety of diseases, which affects its functioning. Urolithiasis is one the third most frequent urological problems among people worldwide [1]. Stone illness influences 2-20% of populace around the world with a predominance rate of 15% in India [2,3]. The majority of Ca-Ox crystallizes in the urinary tract under physiological conditions and is then excreted easily in urine. If the crystals are allowed to remain in the kidney, they will expand and develop into stones, causing damage to the renal epithelial cells and providing a site for the development of a stone [4]. Renal cells become acidic when exposed to elevated concentrations of oxalate ions and/or Ca-Ox crystals under pathological conditions. Harm to renal cells causes changes in cell surface properties, revealing the attachment site for crystal adhesion and/or internalization by renal epithelial cells [5,6]. Various medicinal plants have been used as herbal medicines since they have been documented to have antilithiatic activity [7], and they have been a part of traditional medicine since ancient times. The development of plant-protein dependent medicinal drugs is currently the subject of numerous pharmaceutical industries. These plant proteins could be mass-produced and modified with recombinant DNA technology. Because of the presence of acidic amino acids and/or calcium binding domains (EF Hand motifs), the majority of plant-based antilithiatic proteins known to date are anionic [8].
These acidic amino acids bind to calcium, stopping it from interacting with oxalate and causing renal cell adherence [9]. The present study aimed to assess the bioactivity of anionic anti-calcifying proteins from dried seeds of Macrotyloma uniflorum (Horsegram) on Ca-Oxalate crystals to provide objective support for M. uniflorum's anti-urolithiatic ability. This Fabaceae-family plant is widely used in herbal medicine to treat several ailments. This crop, which belongs to the Fabaceae family, is commonly used in traditional medicine for a variety of cardiovascular disorders. It also has antioxidant, anti-inflammatory, and moderate diuretic properties [10]. M. uniflorum has also been shown to suppress CaOx crystallization and crystal adhesion to renal epithelial cells in vitro [11-13]. The aim of this analysis was to isolate and classify antilithiatic proteins from M. uniflorum seeds, as well as determine their impact on different stages of CaOx inhibition.
Materials and Methods
Plant material
Macrotyloma uniflorum seeds were collected from different locations of Himachal Pradesh (north-western states of India) such as Kullu, Chamba, Rampur, Sundernagar, Dharampur and Palampur. The seeds (HPK-4, HPKM-317, and HPKM-249) from the Palampur region were procured from the Department of Agricultural Biotechnology, CSK, Himachal Pradesh Krishi Vishwavidyalaya H.P., India. All the seeds were authenticated Macrotyloma uniflorum (HP-MU) by Dr. RK Chahota from the Department of Agriculture Biotechnology, CSK Himachal Pradesh Agricultural University, Palampur. The seeds were stored at room temperature.
Protein extraction
Surface-sterilized seeds of M. uniflorum from all locations were soaked in water for 72 h. all the seeds were then crushed and extracted with Tris-buffer [1 gm seed powder was mixed in 3 mL of buffer (pH 8.0) consisting Tris base (0.05 M), polyethylene glycol (1.0%-H 4000), cysteine hydrochloride (0.1%), ascorbic acid (0.1%), citric acid (0.007 M) and β-mercaptoethanol (1 mM)] under cold conditions. The extracted samples were filtered and centrifuged at 12,000 rpm for 20 min at 4°C [Ranjan et al. 2012], and supernatants were assayed for calcium oxalate inhibition potential along with their respective controls. The proteins in all the samples were also estimated by the Bradford method (Bradford 1976).
Electrophoretic techniques
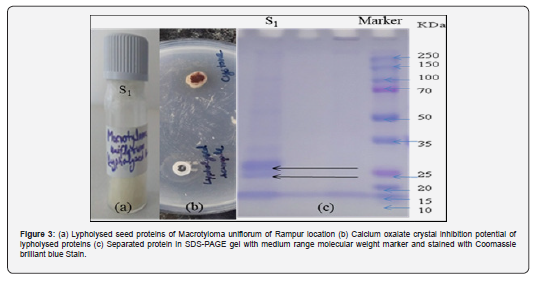
The protein sample showing highest calcium oxalate inhibition activity was then separated by electrophoresis in 12% polyacrylamide gels in the presence of Sodium Dodecyl Sulphate (SDS-PAGE). Extracted and lipolyzed protein samples were added 3:1 to 4X SDS-PAGE sample buffer. The gels were then stained with Coomassie brilliant blue R-250 staining solution. The gels were imaged in a gel documentation system.
Trypsin digestion
A total of 50 μg of proteins from each sample was diluted in 100 mM ammonium bicarbonate to final volume of 100 μl, reduced with 10 mM DTT and alkylated with 55m Miodoacetamide followed by trypsin digestion (Promega, India) for 12-16 h at 37°C. The dried peptide mixture was suspended in 50% trifluoroacetic acid (TFA) containing 0.1% acetonitrile (ACN) buffer. The peptides obtained were mixed with α-cyano-4-hydroxycinnamic acid (HCCA) matrix in 1:1 ratio and resulting 2μL was spotted on the MALDI TOF/TOF ULTRAFLEX III instrument and further analysis was done with FLEX ANALYSIS SOFTWARE for obtaining the Peptide.
Peptide mass fingerprinting
Digested peptide samples were lyophilized (Labconco, USA) and dis- solved in 0.1% triflouro acetic acid /acetonitrile and added to 0.5μl of matrix [α-Cyano-4-hydroxycinnamic acid (Bruker; 20 mg/ ml in 0.1% trifluoroacetic/30% (v/v) ACN (1:2)], dried at room temperature and subjected to MALDI-TOF/TOF proteomics analyzer (UltrafleXtreme TM MS; Bruker Daltonics, Germany). A mass standard starter kit (Bruker Daltonics, Germany) and a standard tryptic BSA di- gest (Bruker Daltonics, Germany) were used for MS and MS/MS cali- b rations. A combined MS and MS/MS were performed using Bio Tools 3.0 software (Bruker Daltonics, Germany). The TOF spectra were recorded in positive ion reflector mode with a mass range from 700 to 3500Da. The two most abundant peptide ions were subjected to fragmentation analysis to determine the peptide sequence. Database search was performed using MASCOT search engine (v.2.1; Matrix Science, London, U.K.). The search parameters in NCBInr were as follows: taxonomy, Viridiaplantae (green plants 25779625 sequences); enzyme, trypsin; fixed modification, carbamidomethyl (C); variable modification, Glu-> pyro-Glu (N-term Q) and oxidation(M); missed cleavage, one; parent ion mass tolerance, 100 ppm; MS/MS mass tolerance, 0.7 Dalton; and MS/MS peak filtering, monoisotropic and M+H+. The identified proteins which met three criteria viz., 1) be among the 5 top hits on the search report, 2) having significant ions scores/sequence coverage or extensive homology (p < 0.05) and 3) proteins matched by a minimum of two peptide sequences were considered as positive identification.
Gene ontology using Blast2Go software
The identified proteins were grouped into functional categories using Blast2GO (v. 2.6.0) [14].
Results and Discussion
MALDI-TOF Mass spectrometric analysis and identification of purified proteins
The crude proteins obtained after extraction of seed samples of Macrotyloma uniflorum from different locations were subjected to MALDI-TOF MS and MASCOT search engine analysis. The mass over charge ratio data obtained from the MALDI-TOF of the peaks A1, B1, B2, C1 matched significantly with nuclear pore anchor, DEAD Box ATP dependent RNA helicase 45, Lon protease homolog 1 and Heat shock protein 90-3, respectively. The amino acid sequence of these respective proteins obtained from MASCOT search was used as an input to search for the presence of active domain using Scan Prosite. Merchant et al., [15] used MS-MALDI and ESI-MS to compare to a Swiss Prot human protein database and a translated human genome database, respectively. They discovered 158 proteins in the stones of five patients. They also discovered large levels of myeloperoxidase and osteopontin, indicating that proteolytic action is involved in stone formation [15] (Table1).

Functional classification
Figure1A depicted the biological and molecular function annotated using Blast2GO. MALDI-TOF-MS identification showed a total of 31 proteins using NCBI green plants. A total of 31 proteins were identified which could be functionally characterized into developmental process, anatomical structure development, tube development and multicellular development. 1-Aminocyclopropane-1-carboxylate synthase (ACC) noticed in Chamba sample. ACC is an enzyme mainly involved in the synthesis of ethylene (Oz, Gulen, & Eris, 2010) and is likely to be part of the sequence of ripening events, with ethylene being a well-recognized trigger for fruit softening and cell wall breakdown, produced during the climacteric [16]. RNA polymerase II is a general transcription factor, present in nucleus and mainly in protein biosynthesis, transcript and transcription regulation. Ribosomal protein L16 (chloroplast) was found in Bharmour sample. Chloroplast r-proteins are encoded within two genetic compartments, the chloroplast and the nucleus and are much conserved structures. Ribosomal protein L16 (chloroplast) protein is mainly involved in mitochondrial translational network.
Genes mainly involved in ribosomal proteins are mainly present in chloroplast genome [17]. Putative tRNA pseudo uridine synthase Pus10-like, partial is metal ion binding protein prominently present in Kullu protein sample which involved in primary miRNA processing and pseudo uridine synthases are responsible for installation of pseudo uridine modification in RNA [18]. Molecular processes involved transporter activity, ion transmembrane transporter activity and carboxylic/ dicarboxylic acid transmembrane activity. Calcium ions (Ca2+) play essential roles in plant growth and development (Figure 1B). Calcium-dependent protein kinase 29-like (CDPK)mainly identified commonly in plants, are a kind of vital regulatory protein deciphering calcium signals triggered by various developmental and environmental stimuli [19]. CDPKs have commonly presented in plants, protists, oomycetes and green algae, but not in animals and fungi [20].
Number of peptides generated in the protein samples of different locations using MALDI-TOF
The protein extracts of seeds of Macrotyloma uniflorum from different locations of Himachal Pradesh were also analyzed for number of peptides generated in MALDI-TOF. The protein sample of Rampur location contained maximum number of peptides followed by Sunder Nagar, Palampur and Kullu. Seeds of Chamba location showed the presence of very few peptides (Figure 2). The kidney stones are known to be composed of mainly calcium oxalate and Macrotyloma uniflorum seed extracts provide a good preface for these stone degradations. The seed protein of Macrotyloma uniflorum with highest calcium oxalate inhibition potential was lipolyzed and then separated through SDS-PAGE technique. The quantification of Macrotyloma uniflorum seed proteins was done with Bradford method and further protein with high oxalate inhibition activity (15µg) was loaded on 12% Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) gel according to Laemmli discontinuous system (Figure 3). In the proteomic study of loaded protein, a total of three major low molecular weight protein bands were observed (~27KDa, ~23.1 and ~17KDa respectively). These results will help in correlating the protein composition and bring us closer to treatment for kidney stone degradation. SDS-PAGE technique can be used to identify genotype in the future, as well as to broaden genetic diversity and relatedness in cultivars, as well as to distinguish mutants from their parent genotype based on seed protein [21].

Summary
In plants, MALDI-TOF MS profiling has been limited to metabolite profiles and has only been documented in a few cases [22]. As a result, no archive containing spectra from protein profiles in plant tissues or organs in physiological or altered states exists to date. However, this technique's use in plant proteomic studies may have a big effect, helping with sample differentiation and/or protein marker discovery in agriculture and industry. The discovery of biological markers is valuable areas of research that can help breeders pick cultivars that are best suited to a variety of biotic and abiotic stresses, as well as different stages of growth. Overall, MALDI-TOF MS profiling is an important tool with many possible applications in plant proteomics, such as protein marker discovery, and can contribute significantly to genetic breeding programs and biotechnology. While efforts must be taken to adapt the technique for various plant tissues, our research has demonstrated that this technology is feasible [23]. Also, seed protein SDS-PAGE may be able to also be used to investigate genetic diversity and relatedness in cultivars at a low cost. Furthermore, the proteomic analysis of the seed protein of M. uniflorum aids in the creation of a protein-based marker that can be used to identify cultivars in the future.




No comments:
Post a Comment