Pharmacology & Clinical Research - Juniper Publishers
Abstract
Objective: The objective of this experiment was to investigate the inhibitory activity of targeted quassinoids from the Simarouba glauca employing molecular docking & ADME analysis against selected ligands. Methods: The 3D structures of ligands were fetched from Pubchem database and subjected to bioinformatics tools such as ADME analysis and Molecular docking simulations to predict the new leads for treatment of HIV-1 infections. Results: The comparison of molecular docking score exposed that the selected compounds showed better binding affinity towards the known co-crystals. And ADME properties and Lipinski parameters for drug analysis making them potentially momentous agents for Bio-activities. And finally, the results exposed or showed that these selected compounds from Simarouba glauca are expected to be beneficial & effective in HIV-1 infections.
Keywords: Simarouba glauca; ADME analysis; Lipinski filter analysis; Quassinoids; Anti – HIV1
Introduction
Simarouba glauca has a great history of use in traditional or herbal medicines across the world and mainly the leaf & bark extract of Simarouba is best known for its various pharmacological activities like antihelmenthic, antipyretic, anticancerous [1], antiulcer [2], antiamoebic, antiprotozoal [3] and many more. The plant includes mainly: ailanthinone, benzoquinone, canthin, dehydroglaucarubinone, glaucarubine, glaucarubolone, glaucarubinone, holacanthone, melianone, simarolide, sitosterol, & tirucalla. After discovery of bruceantin (a quassinoid), which is responsible for antileukemic activity, these phyto-molecules gained very much attention [4]. Around 150 quassinoids have been isolated depends on their chemical structures [5]. Considering the future in developing anti-HIV agents with more effective & less harmful compounds, indigenous quassinoids represent a resource of small molecules. The selected quassinoids, some were selected (ailanthinone, benzoquinone, canthin, dehydroglaucarubinone, glaucarubine, glaucarubolone, glaucarubinone, holacanthone, melianone, simaroubidin, simarolide, simarubin, simarubolide, sitosterol, & tirucalla) based on PASS prediction server [6]. The given ligands were predicted to collaborate with both CXCR4 & CCR5. Both of these molecules (CXCR4 & CCR5) are chemokine receptors which are present in different types of cells like macrophages, monocytes, T-cells as well as acts as co-receptor for HIV-1 in these cells. CCR5 was first isolated as a working GPCR which is acted against 3 CC chemokine [7,8]. Among the many CC chemokine that have been noted to bind, CCR5 show the most suppressive activities in HIV-1 infection assays [9] and CCR5 also acts as a potential enhancer despite than an inhibitor of HIV-1 replication [10]. CXCR4 was initially identified as leukocyte-derived seven-transmembrane domain receptor [11-15] but did not get so much response until its isolation as a co-receptor for HIV-1 [16]. Zidovudine, didanosine, zalcitabine, stavudine, lamivudine, abacavir, and emtricitabine are potent selective inhibitors ligand and clinically approved by the US and European Union and is nowadays recommended for cure of HIV-1 patients. In the present study, we tried on comparative analysis of the selected quassinoids from the Simarouba glauca plant with the known drug and carried out the ADME analysis and molecular docking which investigated the drug likeness of the ligands
Methodology
Preparation of Ligand
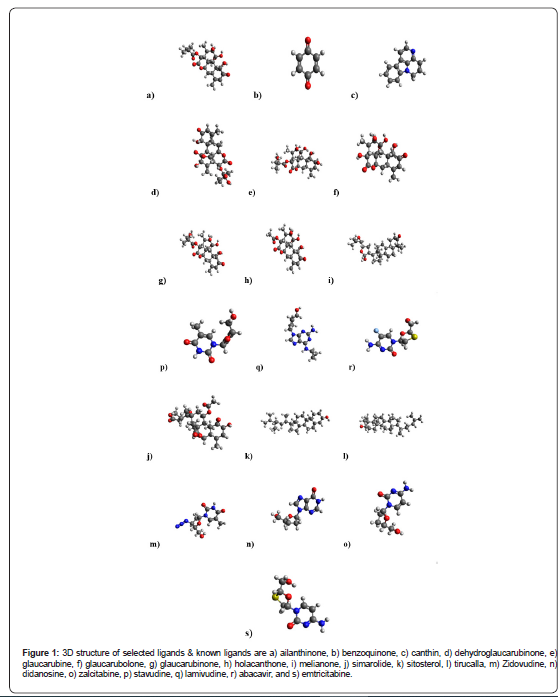
Ligands bind to the protein’s binding sites and the SDF (structure data format) files were obtained of all compounds from Pubchem database [17] & analyzed by Marvin view [18]. And then compounds were transformed into 3D structure using PyMol version 1.3 [19]. And physico-chemical properties of the ligands are estimated by Pubchem open chemistry database.
Investigation of drug likeness of the ligands
Lipinski filter were used for drug likeness prediction and according to orally active drug must fulfill some criteria for drug likeness mainly cLogP, molecular mass, hydrogen donor & acceptor, & molar refractive index [20]. And all of these properties were investigated by SWISS ADME, which is noted as a conventional drug discovery tool [21] as well as brain access & gastrointestinal absorption are 2 pharmacokinetic behaviors to measure the various levels of the drug discovery mechanisms. And the Brain or Intestinal EstimateD permeation method (BOILEDEgg) is proposed as an exact predictive model [22].
Molecular Docking Analysis
The objective of the molecular docking is to predict the active binding modes of ligands & orientation of molecules with respect to binding sites. According to this docking, the affinity score gives the ranking of all the binding pores of molecule inside the catalytic site of an enzyme is being done and the targeted compounds were selected for minimization using Ligprep module of Schrodinger. In molecular docking, the receptor grid was analyzed keeping (zidovudine, didanosine, zalcitabine, stavudine, lamivudine, abacavir, and emtricitabine) on (ailanthinone, benzoquinone, canthin, dehydroglaucarubinone, glaucarubine, glaucarubolone, glaucarubinone, holacanthone, melianone, simaroubidin, simarolide, simarubin, simarubolide, sitosterol, & tirucalla) at the centre of grid with 20 A edges bearing catalytic sites. Moreover, the molecular docking was performed for given ligands against Protein Data Bank ID using Glide XP 5.8 program [23-25]. After docking, the lowest energy or highest score given by Glide program was selected as the most probable binding pose of top compound.
Results & Discussion
Ligands
(Figure 1 & Table 1) The known drugs exposed polar surface area is around 77 Ų- 135 Ų, whereas many selected compounds having less polar surface area like benzoquinone (mass – 108.09 g/mol) followed by canthin, holacanthone, & melianone.
Analysis of Drug Likeness
After completion of Lipinski filter analysis which exposed the rigidity of all compound to be remembered for structure-based drug design and also listed out the compound’s properties with similar to their usage using ADME analysis (Table 2).
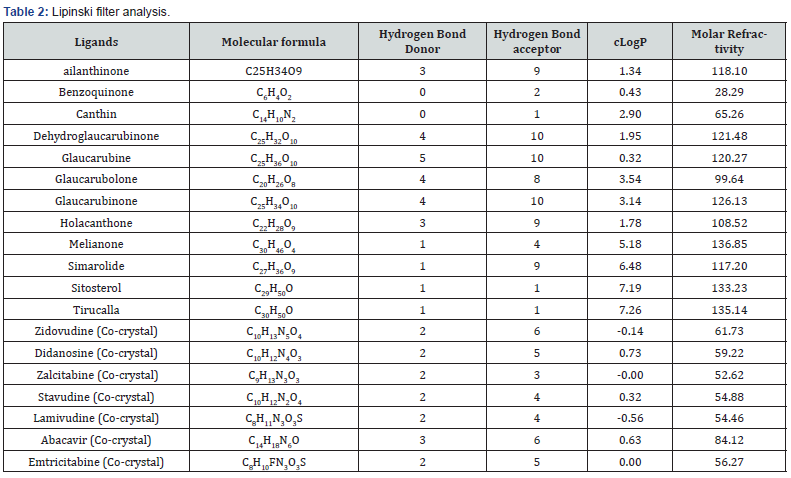
Criteria: log P≤5.0, molecular weight in the range of 150–500, H-bond donor’s ≤5, and H-bond acceptors ≤10. The result come from the above table indicates that the ligands selected were noted to be in acceptable range defined for human use which shows their potential drug like property (Table 3).
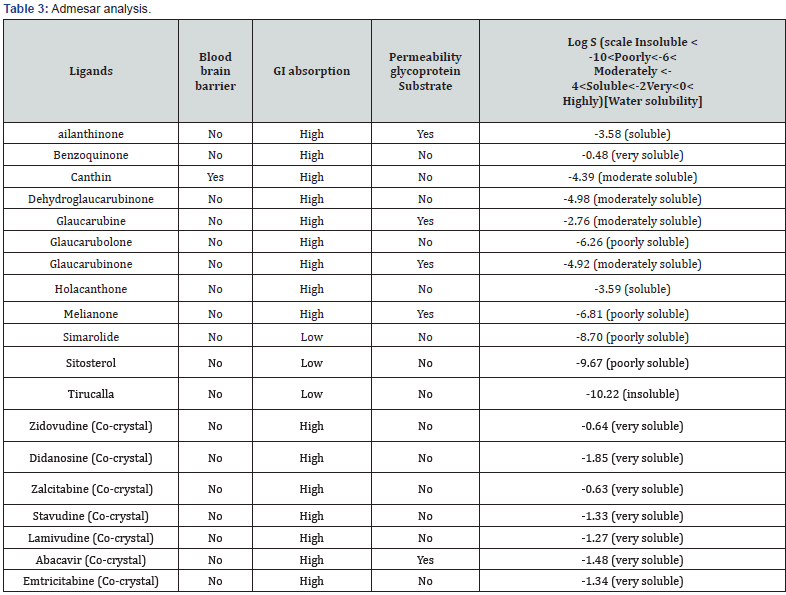
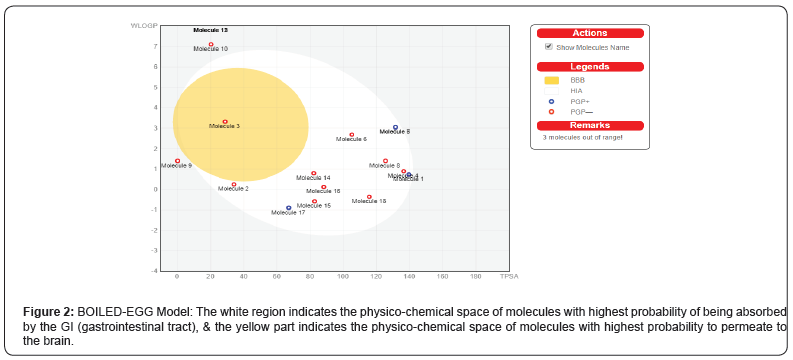
Brain or Intestinal Estimated permeation method (BOILED-EGG)
The prediction also reveals that Benzoquinone & Holacanthone has high GI absorption followed by known drugs Zidovudine, didanosine, zalcitabine, stavudine, lamivudine, abacavir, and emtricitabine which is then followed by canthin, Dehydroglaucarubinone, Glaucarubolone have low GI absorption (Figure 2).
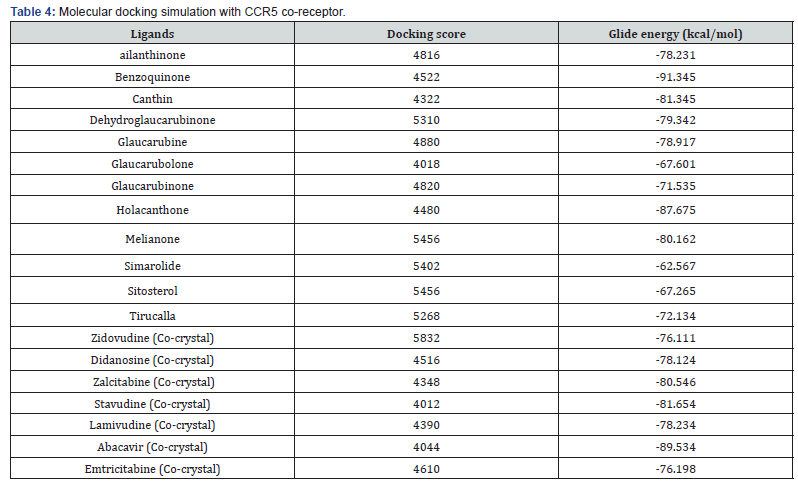
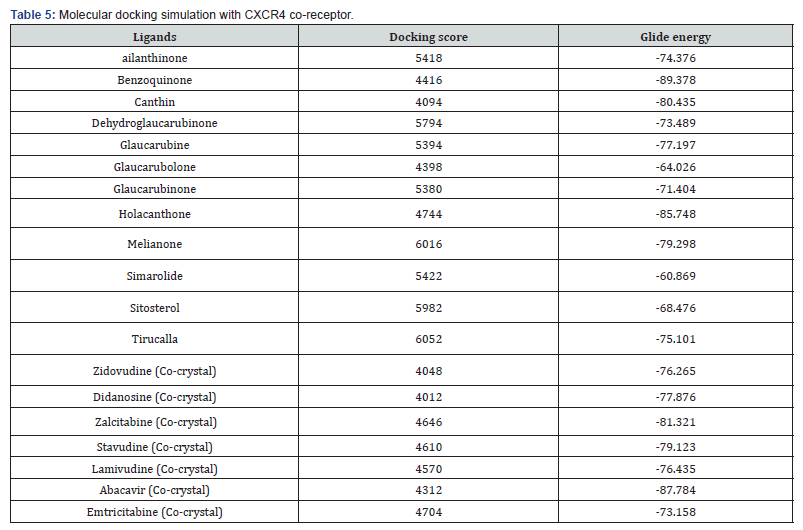
Molecular Docking Results
The results obtained from molecular docking of selected compounds with CCR5 &CXCR4 receptors shows that, most of the compounds are exposing better docking score and energy than that known drugs Zidovudine, didanosine, zalcitabine, stavudine, lamivudine, abacavir, and emtricitabine which predicts that the compounds taken have the better binding affinity towards the receptor than the co-crystal. And this prediction indicates us to believe that the compounds will be suitable or proper for treatment of HIV-1 infection (Tables 4 & 5). In selected compounds, benzoquinone showed best glide energy in both receptors (-91.34 & -89.37) followed by holacanthone (-87.67 & -85.74), which is far better than the known drugs score and glide energy.
Conclusion
The in-silico studies of the selected quassinoid compounds from the Simarouba glauca exposed favorable outcomes & the chosen compounds showed better docking score and adme analysis properties than known drugs. Based on ADME prediction Benzoquinone & Holacanthone has high absorption & very soluble compared to co-crystal drugs. The Benzoquinone selected from Simarouba glauca are non-toxic. Hence, it has been identified and predicted that all compounds may possibly serve as new leads for the cure of HIV-1 infections and this results might be in future used in in vivoo studies to test their effects on the cure of HIV-1 infections.
To Know more about Pharmacology & Clinical Research
Click here: https://juniperpublishers.com/index.php





Hi, this is Arunprakash. I like the way the article is explained. The content is very good. If any of the engineering students who are looking for a matlab projects on image processing I found this site and they are providing the best service to the engineering students regarding the projects matlab projects on image processing
ReplyDelete