Trends in Technical & Scientific Research
Abstract
Nature has its own way of synthesizing molecules and
materials with an array of bio-catalysts known as enzymes. Enzymes
comprise of a combination of proteins which in turn are the building
blocks of life. With just a backbone combination of Carbon, Hydrogen,
Oxygen and Nitrogen, proteins can be perceived as delightfully
intelligent and adaptive. Over billions of years, nature have provided a
wide range of survival questions in front of these molecules and
somehow, they have evolved and survived, as proved by Darwin. This
adaptive nature of proteins has been exploited and successful evolution
of enzymes is possible towards a desired interest on a laboratory scale
using nature’s machinery. Thus, mimicking evolution on a much shorter
time-scale. In this article, we address the strategies involving the
idea of evolution in a laboratory directed to fulfil our chemical needs.
Also, how with time, the strategies have evolved covering up the
loop-holes of previous generations. The advent of polymerase chain
reactions made combinatorial libraries creation easy and then screen
towards a fitness test. With time we learnt, a combination of evolution
with rational designing creates shorter and smarter libraries that have a
strong ‘impact factor’ upon the desired goal and reduce
false-positives. However, while controlling biological systems,
scientists are manipulating the factories that actually make the
molecules and materials into newer forms that are not natural. Nature,
out-does scientists via evolution. Through survival of the fittest,
evolution collects all the beneficial mutations through multiple
iterated genetic diversification and screening.
Keywords:
Directed evolution; Protein engineering; Combinatorial libraries;
Mutagenesis; Enzymes; Promiscuous ancestral proteins; Escherichia coli
metabolism; Chemical origins; Thermoanaerobacter brockii alcohol
dehydrogenase; X-ray crystallographic structures
Introduction
Enzymes are thought to be highly specific biological
catalysts having a very discreet range of target molecules or substrates
[1, 2]. The evolution of enzymes has been occurring under different
selection pressures of nature from promiscuous ancestral proteins. A
genomic analysis in Escherichia coli metabolism reveals that 37% of its
enzymes have a wide variety of substrate range. However, they are found
to catalyze 65% of the known metabolic reactions [3]. The evolution of
enzymes in response to environmental demands is the key to the overall
organism’s fitness in the nature. This is manifested in the inherent
promiscuity of many enzymes. Although, we are thankful to the
‘specialist’ nature of the enzyme molecule, we also do question, how
much further can the ‘generalist’ nature be pushed in-vitro or in-vivo
so that an enzyme can be evolved more. In the 3-D structure analysis,
the active site of an enzyme is the engine, that performs the turn-over
of a substrate into the product (catalytic machinery) and more
specifically amino-acid residues have been identified from the X-ray
crystallographic structures that are key players in the process.
Textbook definition of enzymatic activity reveals that the work of the
enzyme is broadly related to the binding of the substrate into the
active site, followed by a possible conformation change, then the
catalytic activity happening in the core of the enzyme and finally, the
product leaving the enzyme-product complex. To explain the evolution of
enzymatic catalytic activities, scientists have used models of chemical
origins to propose the evolutionary initiation of the diversity. One
such model demonstrates via chemical models and assumes key mutations in
the active site that can lead towards enhanced specificities or
sometimes enhanced specificities but with compromised efficiency. Then
it proposes that designing a realistic trade-off constraint is possible
with mutation combinations where the evolution can be directed towards
both enhanced specificity and efficiency [4].
To improve the characteristics of an enzyme
structural bioinformatics has been extensively used and with de-novo
approaches enzymes have designed for enhanced specificity and
functionality. Evolution has been a brute force that pushes the enzymes
to evolve and then select the survivors based on their functionality.
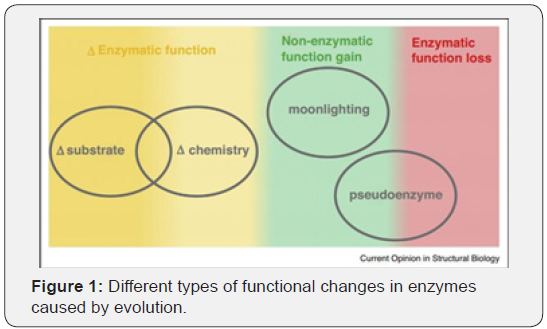
From Figure 1 it is evident that the evolution not only results in
various changes in chemistry and substrates but are also supplemented
with rarer gain or
loss in the function in the form of moonlighting and pseudoenzymes
respectively [5]. Due to the innate promiscuity, many
enzymes do show off-target activity even when the natural
selection pressure ceases to exist over evolutionary time. At
that point of saturation, further catalytic/specificity changes
do not improve the fitness in its highly conserved active site.
The phenomenon was demonstrated by -lactam antibiotic
resistance in bacterial populations over many generations.
The promiscuity can also vary between orthologous enzymes
which originate from the same family [5-7]. An approach of
rational design relying on somewhat of a thorough knowledge
of the protein structure that exists for many years is called
directed evolution [8,9]. It is a popular technique that has been
used to evolve a particular enzyme towards a user-defined
goal. The ‘user defined’ goals narrows down the chances to
produce the false-positives in comparison to natural selections.
Also, this gives an opportunity to mimic natural evolution
in a laboratory pilot scale operation. The limitations of this
technique however is, screening may be needed for enormous
random mutagenesis libraries which in-turn needs a robust
high-throughput assay. A combinatorial approach has also
been implied to create ‘focused’ libraries that will concentrate
on regions of the enzyme active site that has been predicted
by rational design to produce beneficial mutations and reduce
the number of mutants to screen by a significant number.
However, it must also be mentioned that our understanding of
the link between the genetic sequence and function lags well
behind our desire for designing new enzymes. Which makes
predictions of performance-enhancing mutations, extremely
challenging.
In the early 90s the most user-defined properties that
were targeted for evolution included thermostability, nonnative
substrates or activity in harsh organic solvents and
after rounds of screening it was sometimes revealed that
beneficial mutations could be nowhere near the active site
of the enzyme [5,6]. Hence, it would be a hitting a deadended
loop to just look for the key in the active site of the
enzyme. The power of a combinatorial library is enormous.
With a stable high-throughput assay it is possible to analyze
millions of mutant proteins in one day. In some other cases
however, the number can be less than a few hundred per day.
So, development of smarter libraries is very important in this
aspect. Using site-directed mutagenesis and site-directed
saturation mutagenesis techniques, identification and
construction of a ‘mutable landscape’ has been done in case
of cytochrome P450 evolution [10-12]. This has been proved
to be a very effective technique recently to tackle the very
important challenge of the sequence-function relationship
in case of directed evolution. The libraries thus generated
on mutability landscapes can be used to engineer any fitness
trait of user-defined interest [6,7,13]. We are entering into an
era where the ‘user-defined’ interest has changed a lot. The
demand for synthetic compounds has been growing and ‘green
synthesis’ has been one of the most valued tools in the 21st
century that uses enzymes as biocatalysts and minimizes the
production of non-degradable bye-products and pollutants. So,
directed evolution of stereo- or regio-selective enzymes that
can behave as biocatalysts in asymmetric transformations is
of particular interests to bio-organic chemists. Towards this
goal, raised the need to develop the small and smart libraries.
And hence forth different strategies including triple-code
saturation mutagenesis (TCSM) at multiresidue sites of the
Thermoanaerobacter brockii alcohol dehydrogenase by using
distinct reduced amino acid alphabets and with requirement
minimal screening have been recently reported [13-15]. So
not only the physical labor has been greatly reduced, but also
chances of false-postive results and codon-degeneracy can be
minimized under this protocol [13].
Discussion
To push the promiscuous nature inherent in enzymes has
been what scientists been brain-storming over the decades.
The further challenges are to create enzymes for reactions
that will let them solve problems that neither synthetic
organic chemistry nor biological chemistry can address. Being
macromolecular catalysts, it would be easier for enzymes
to stabilize transition-states which otherwise would be
inaccessible to chemical small molecule catalysts as they would
likely compete with other lower-energy reaction pathways.
Such was reported from cytochrome P450 from Labrenzia
arregata [16]. Strategies and efforts are currently being
invested to design enzymes with artificial cofactors as well.
However, designed enzymes are yet to have the sophistication
of nature’s products and struggles remain with other co-factors
and prosthetic groups that are currently being addressed.
Conclusion
Over time, nature has been the best template to
mankind.
In our case, it has been the best chemistry teacher, solving the
very challenges of existence of life over billions of years through a
range of astonishing conditions. This has been possible only
at the molecular level because of the enzymes which act as the
fundamental bio-molecules. These systems are good models
for a sustainable chemical industry or energy industry that
uses renewable natural resources and recycles almost all of
it without causing significant damage to the environment.
It is not far where we stand today that DNA programmable
organisms can produce many of our desired chemical needs.
To Know More About Trend in Technical & Scientific Research Please click on: https://juniperpublishers.com/ttsr/
To Know More About Open Access Journals Please click on: ttps://juniperpublishers.com/index.php




No comments:
Post a Comment