GastroenterologyOpen Access Journals Juniper Publishers
Authored by Abdulghani Mohamed Alsamarai
Abstract
Background: Genetic variations of the infectious agents play an important role on the influence of natural disease severity, response to treatment and morbidity.
Aim: To clarify if there are genetic variations in H. pylori clinical isolates from cases with gastritis, peptic ulcer and carcinoma of stomach.
Materials and Methods:Eight PCR product sample were sent for sequence analysis of H.pylori isolated from biopsy of stomach and 20μl (10pmol) from the Forward primer sequence is 5′-GGTTCAAATCGGCTCACACT-3′ Reverse primer sequence is 5′-CTCCTTAATTGTTTTTACATAG-3′ They are made by primer 3PLUS software. The result of the sequence analysis was analyzed by blastn in the National Centre for Biotechnology Information (NCBI) and many software used to detect polymorphism in H. pylori.
Results: The present study H.pylori isolates were separated on the phylogenetic tree and belong to a population cluster different from previously reported H.pylori sequences. H.pylori variant 1 which was isolated from Iraqi patients with gastric cancer was most closely to strain Helicobacter pylori R037c isolated in Canada. While variants 2 and 3, which were isolated from peptic ulcer and variant 4, which was isolated from patient with gastritis were more closely to Helicobacter pylori 26695 isolated from patient with gastritis in UK. The present study data suggest that sub lineages evolution within the Helicobacter pylori species may be associated with the development of less or more pathogenic potential. In addition, the relation of the 4 variants to the American [Hp R037c] and European [Hp 26695] suggest that these variants diverged from American and European lineage ancestors as reported previously for other geographical areas. This study is the first one in Iraq that sequenced H.pylori isolates from patients with gastritis, peptic ulcer and gastric cancer and the gene diversity of the bacteria may be used as biomarker that may be used for follow up and prognosis in the above disease for prevention of development of cancer. Therefore, called GhaFa.1. Iraq for first isolate, GhaFa.2. Iraq for second isolate, GhaFa.3. Iraq for third isolate and GhaFa.4. Iraq for fourth isolate.
Conclusion: This study indicated that genetic variation in H. pylori was correlated to gastritis severity and was more in gastric cancer cases as compared to gastritis and peptic ulcer.
Keywords: H. pylori; Gastritis; Peptic ulcer; Carcinoma of stomach; Genetic variation; Sequencing
Abbrevations: NCBI: National Center for Biotechnology Information; PCR: Polymerase Chain Reaction; SNP: Single Nucleotide Polymorphism
Introduction
H. pylori infections are worldwide and affected many systems beyond the stomach [1]. However, the disease prevalence varied between different studies [2]. The infection was with a range of 28.3% to 100% in general population and it was more in subjects with gastritis and gastric carcinoma [2]. H. pylori infections was linked with gastric carcinoma [3], however, not all H. pylori infected individuals will develop carcinoma of the stomach. Thus, development of gastric carcinoma and its association with H. pylori infections may interfere with risk factors which lead to potentiate cancer in certain subjects and not the others [3,4]. Previous studies reported abnormality in biomarkers in individuals with gastritis, peptic ulcer and gastric carcinoma, and the abnormality was more prominent in subject with cancer of stomach [5,6].
Genetic variations of the infectious agents play an important role on the influence of natural disease severity, response to treatment and morbidity [7,8]. Thus, this study was conducted to clarify if there are genetic variations in H. pylori clinical isolates from cases with gastritis, peptic ulcer and carcinoma of stomach.
Materials and Methods
Eight PCR product sample were sent for sequence analysis of H.pylori isolated from biopsy of stomach and 20μl (10pmol) from the Forward primer sequence is 5′-GGTTCAAATCGGCTCACACT-3′ Reverse primer sequence is 5′-CTCCTTAATTGTTTTTACATAG-3′ they are made by primer 3PLUS software. The result of the sequence analysis was analyzed by blastn in the National Centre for Biotechnology Information (NCBI). Online at (http:// www.ncbi.nlm.nil.gov) and many software used to detect polymorphism in H. pylori.
Results and Discussion
The result shown in Figure 1 A-C indicated that a yield of single band of the desired product with a molecular weight of 301bp for urease gene of H. pylori was obtained from 8 samples. Eight PCR product sample were sent for sequence analysis of H.pylori isolated from biopsy of stomach and 20μl (10pmol) from the Forward primer sequence is 5′-GGTTCAAATCGGCTCACACT-3′ Reverse primer sequence is 5′-CTCCTTAATTGTTTTTACATAG-3′
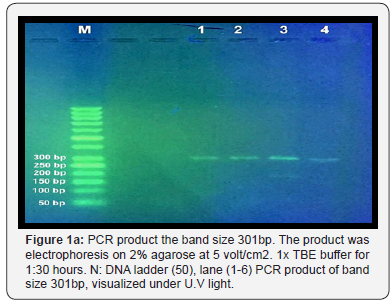
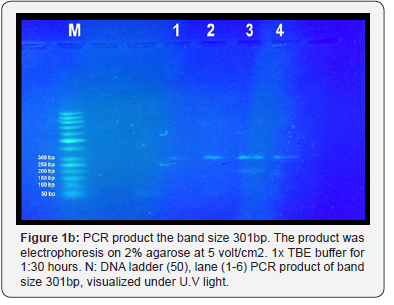
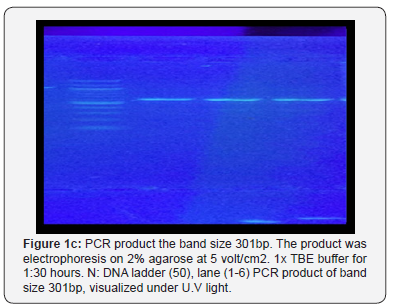
They are made by primer 3PLUS software. The result of the sequence analysis was analyzed by blastn in the National Centre for Biotechnology Information (NCBI). Online at (http:// www.ncbi.nlm.nil.gov) and many software used to detect polymorphism in H. pylori found variation include Transversion (refers to the substitution of a (two ring) purine or a (one ring) pyrimidine and transition a (point mutation) that changes a purine to another purine or pyrimidine to another pyrimidine
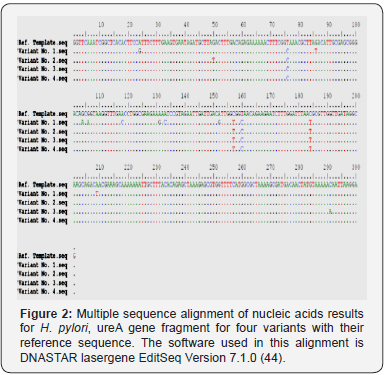
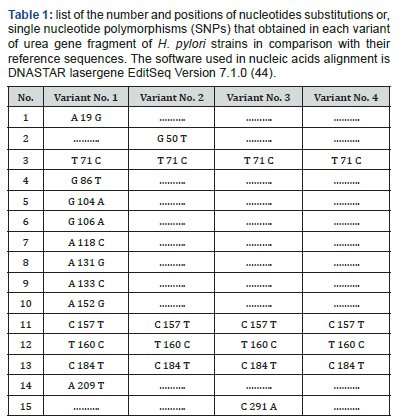
The polymorphisms of Helicobacter urease subunit alpha gene were detected by PCR / direct DNA sequencing method. Undoubtedly, too small sized of the studied groups is still a shortcoming of our research. Nonetheless, the observed sequencing variations nucleic acids indicate the detection of four different genetic variations with variable number of single nucleotide polymorphism [SNPs] (Figure 2). Sequencing results observed that variant No. 1 possesses very high SNPs variability compared with the reference as well as other variants nucleic acid sequences. The pattern and nature of each SNP that detected by sequencing may indicate that these SNPs are novel in their positions and consequences. The variant No. 1 has 13 SNPs (A 19 G, T 71 C, G 86 T, G 104 A, G 106 A, A 118 C, A 131 G, A 133 C, A 152 G, C 157 T, T 160 C, C 184 T, and A 209 T), and only four (T 71 C, C 157 T, T 160 C, and C 184 T) are in common with other three variants (Table 1). The variant No. 2 has 5 SNPs (G 50 T, T 71 C, C 157 T, T 160 C, and C 184 T), and four (T 71 C, C 157 T, T 160 C, and C 184 T) of them are in common with other three variants. The variant No. 3 has 5 SNPs (T 71 C, C 157 T, T 160 C, C 184 T, and C 291A), and four (T 71 C, C 157 T, T 160 C, and C 184 T) of them are in common with other three variants. The variant No. 4 has 4 SNPs (T 71 C, C 157 T, T 160 C, and C 184 T), and all of them are in common with other three variants, and no one unique SNP was identified.
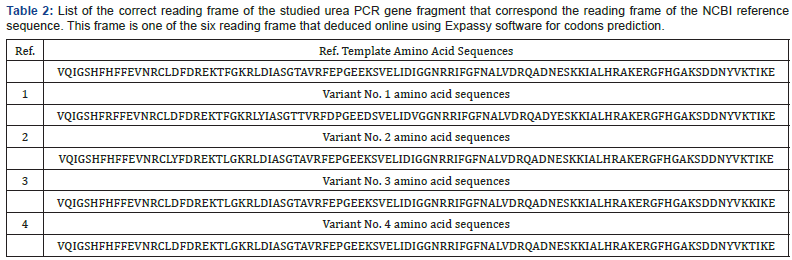
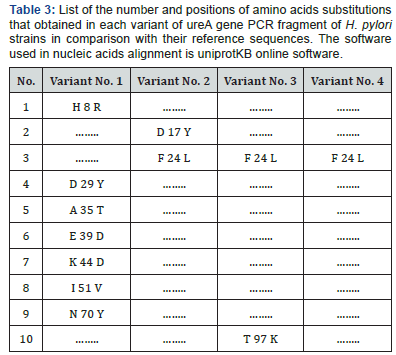
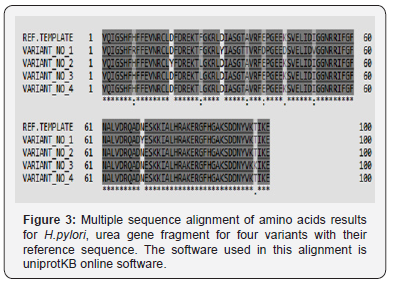
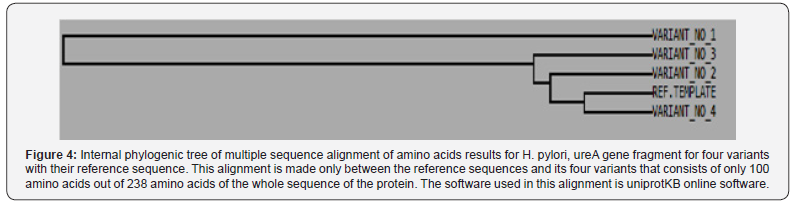
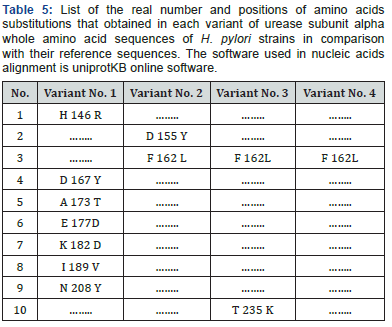
Since multiple reading frames can be extracted from each amino acid sequence, special software were used to predict the correct reading frame that suits the original reference reading frame in urease subunit alpha (Table 2). Once these suitable open reading frames were constructed, they compared with each other using uniprotKB software to see the final effect of the observed SNPs on the primary structure of proteins fragments Figure 3. Moreover, other data that found the high variability of urea gene isn’t only stated in the levels of DNA, but it extends into the proteins’ level too. According to Figure 3, it was found that variant No. 1 has seven amino acid changes (H 8 R, D 29 Y, A 35 T, E 39 D, K 44 D, I 51 V, and N 70 Y), and all of them aren’t in common with the other three variants (Table 3). Variant No. 2 has two amino acids changes (D 17 Y, and F 24 L), and only one of them (F 24 L) is in common with the other three variants. Variant No. 3 has two amino acids changes (F 24 L, and T 97 K), and only one of them (F 24 L) is in common with the other three variants. Whereas variant No. 4 has only one amino acid change (F 24 L), which is in common with the other three variants. According to previously mentioned data, it was found that the variant No. 1 has distinct and significant variations in comparison and the reference, as well as other three amino acids sequences of the other three variants Figure 4. However, to get a complete idea about the extent and nature of these amino acid’s changes, the whole sequence of urease subunit alpha was retrieved, and the accurate position of PCR products were localized. Table 4. Hence, the real positions of these changes were relocalized Table 5 and introduced in the whole sequence of this protein. Accordingly, only ten amino acid changes were represented in all of these four observed variants Table 6. Seven of non-synonymous mutations were represented in variant No. 1 (H 146 R, D 167 Y, A 173 T, E 177 D, K 182 D, I 189 V, and N 208 Y).

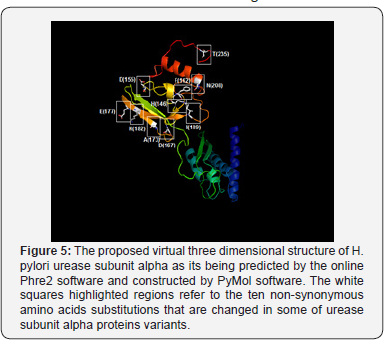
The non-synonym mutations that exist in variant No. 1 was shown seven amino acids changes. The conversions of histidine into argenine, lysine into aspartate, glutamate into aspartate, all polar charged amino acids, and isoleucine into valine, both hydrophobic aliphatic amino acids (in the positions 146, 177, 182, and 189 respectively), may not have a drastic effect on the protein structure since all of these converted amino acids residues have the same functional units Figure 5 & 6.
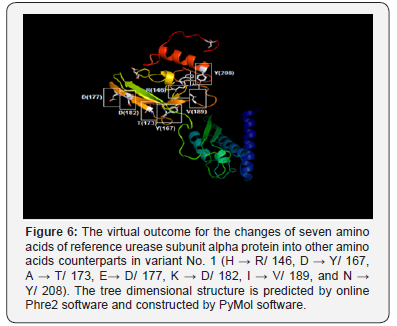
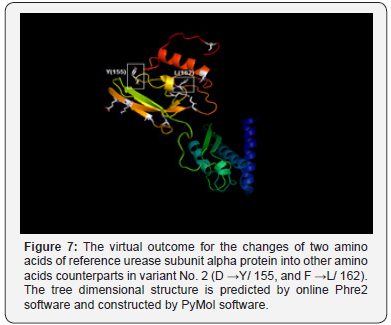
Whereas, the conversion of aspartic acid, the polar charged amino acid into tyrosine, the hydrophobic aromatic amino acid, alanine, the hydrophobic aliphatic amino acid into threonine, the polar uncharged amino acid, and asparagine, the polar uncharged amino acid, into tyrosine, the hydrophobic aromatic amino acid (in positions 167, 173, and 208 respectively) have potential drastic effect of on the final manifestation of the protein structure since all of these converted amino acids residues have different functional units Figure 6. The non-synonym mutations that exist in variant No. 2 was shown two amino acids changes. The conversions of aspartate, the polar charged amino acid into tyrosine, the hydrophobic aromatic amino acid, and the conversion of phenylalanine, the hydrophobic aromatic amino acid, into leucine, the hydrophobic aliphatic amino acid (in positions 155 and 162) may have dramatic effect on protein three-dimensional structure and function Figure 7..
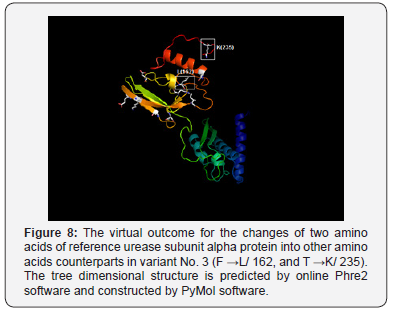
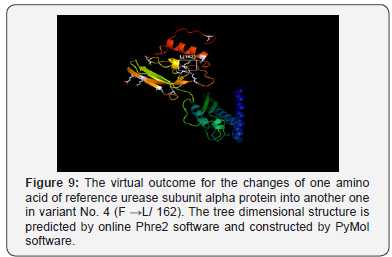
As well, the non-synonym mutations that exist in variant No. 3 was shown two amino acids changes. In addition to phenylalanine / leucine conversion that observed in variant No. 2, another conversion was observed from threonine, the polar uncharged amino acid into lysine, the polar charged amino acid (in position 235) These two changes may have dramatic effect on the three-dimensional structure too Figure 8. In variant No. 4, only one amino acid conversion was observed, which has been observed in both variant No. 2 and variant No. 3. Figure 9. It was obviously found in this study that variant No. 1 is genetically distinct from all other three observed variants as well as the reference sequences Figure 10. Thus, the variant No. 1 is highly polymorphic and may be regarded as a mutational hotspot, or the high variable site, leading to ligand diversity. The high genetic dissimilarity that we observed in this studied variant in comparison with its reference breed sequences might be due to several factors, such as the type, location, or other factors since the nature of polymorphisms were potentially highly dependable on several factors in the determination of its final genetic polymorphism.
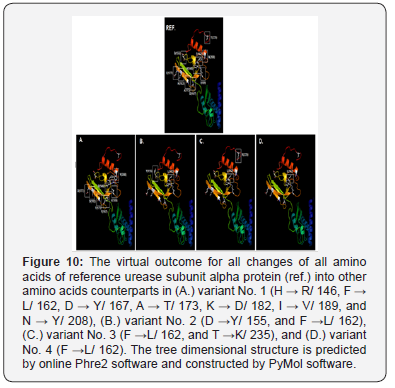
In addition to the closeness of the amino acid’s sequences of variants No. 2, 3, and 4, the phylogenetic tree that we construct has shown that the variant No. 1 has the most far genetic distance compared with the other observed three variants of this study as it was presented through several topological frames Figure 11- 13.
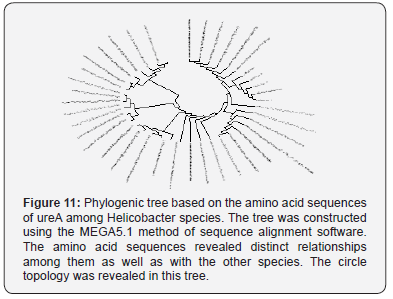
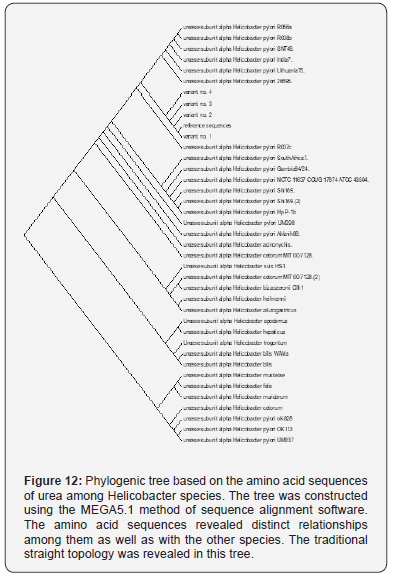
However, the observed variants of the current study are provided a beneficial tool for the geneticists and microbiologists to identify the unknown SNPs that may be useful for establishing a possible association with the genealogical relationships among these variations. The present study indicated that H.pylori isolated from patients with gastric cancer [variant 1] was highly polymorphic and was genetically distinct from H.pylori isolated from peptic ulcer [variants 2 and 3], and from patients with gastritis [variant 4]. These genetic diversity in our isolates was in line of previous studies performed in other geographical areas [8]. In addition, the variability was not confined to DNA only, but it extend to involve proteins and these variability may attribute to evolution of new H.pylori strains with high ability for gastric cancer induction. Cover [8], reported that there was presence of certain proteins in H.pylori with higher gastric cancer risk, which were absent in isolates with low gastric cancer risk. Thus genetic diversity and protein variability in H.pyloriisolates may influence the incidence of gastric cancer and development of gastrointestinal diseases.
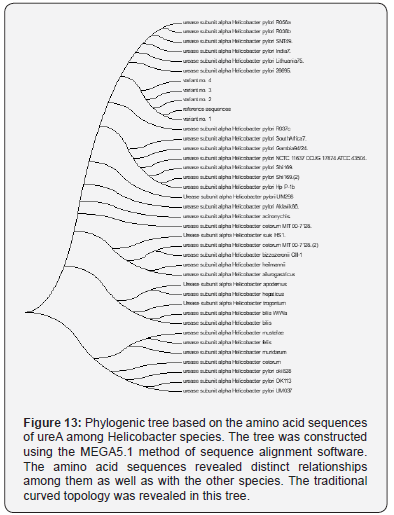
The present study H.pylori isolates were separated on the phylogenetic tree and belong to a population cluster different from previously reported H.pylori sequences [9-12]. H.pylori variant 1 which was isolated from Iraqi patients with gastric cancer was most closely to strain Helicobacter pylori R037c isolated in Canada [11]. While variants 2 and 3, which were isolated from peptic ulcer and variant 4, which was isolated from patient with gastritis were more closely to Helicobacter pylori 26695 isolated from patient with gastritis in UK [9]. The present study data suggest that sub lineages evolution within the Helicobacter pylori species may be associated with the development of less or more pathogenic potential. In addition, the relation of the 4 variants to the American [Hp R037c] and European [Hp 26695] suggest that these variants diverged from American and European lineage ancestors as reported previously for other geographical areas [11,13].
This study is the first one in Iraq that sequenced H.pylori isolates from patients with gastritis, peptic ulcer and gastric cancer and the gene diversity of the bacteria may be used as biomarker that may be used for follow up and prognosis in the above disease for prevention of development of cancer. Therefore, called GhaFa.1. Iraq for first isolate, GhaFa.2. Iraq for second isolate, GhaFa.3. Iraq for third isolate and GhaFa.4. Iraq for fourth isolate.
Conclusion
This study indicated that genetic variation in H. pylori was correlated to gastritis severity and was more in gastric cancer cases as compared to gastritis and peptic ulcer. This finding suggest that organism gene diversity influence the outcome and prognosis of H. pylori infections.
Ethics Approval and Consent to Participate
The research protocol was approved by Tikrit University College of Science Ethical Committee and informed consent was taken from each participant before their enrolment in the study.
Consent for Publication
All the authors were agreed for manuscript publication
Authors Contributions
All the authors were contributed in the study design, samples and data collection, investigational work, data analysis, and manuscript writing.
To
Know More About Gastroenterology Open Access Journals Please Click on:
https://juniperpublishers.com/argh/index.php
To Know More About Open
Access Journals Publishers Please Click on: Juniper
Publishers




No comments:
Post a Comment