Journal ofMedical Microbiology and Diagnosis-Biotechnology Open Access Journals Juniper Publishers
Abstract
The ascomycete Fusarium oxysporum f. sp. cubense is the causal agent of the vascular wilt of banana plants, also known as Panama Disease. Recently, a novel race of this pathogen, known as Fusarium oxysporum f. sp. cubense Tropical Race 4 is considered a devastating factor that seriously endangers the global production of bananas and plantains. Comparative genomics reveals that the genome Tropical Race 4 harbors a higher number of secreted protein genes such as the SECRETED IN XYLEM that are well known virulence factors of Fusarium oxysporum. Since the species Fusarium oxysporum does not present a sexual cycle, alternative mechanisms such as anastomosis might enable horizontal gene transfer for acquiring virulence genes that gave rise to the Tropical Race 4. Conditionally-dispensable chromosomes carrying virulence genes and transposable elements have been identified in virulent strains, and they have to be considered as part of the evolutionary history of this pathogen. The identification of some of those virulence genes in Fusarium oxysporum f. sp. cubense suggests that accessory chromosomes reliably exist in this formae specialis, but in order to be functional, they have to carry their own transcriptional regulators that ensure their autonomous regulation, and also present a gene architecture compatible to the transcriptional machinery that regulates the expression of the core genome. Hence, horizontal gene transfer might be a driving force that spreads virulence factors among relative fungi to produce novel races of banana pathogens.
Keywords: Fusarium oxysporum f. sp. cubense ; Wilt; Secreted in xylem; Banana plants; Panama disease; Pathogen; Global production; Tropical race; Chromosomes; Architecture; Horizontal gene transfer; Production; rhizome; Tropical race; Phytosanitary policies; Vegetative compatibility; Asparagus; Musa acuminata; X Musa balbisiana.; Cabbage
Abbreviations: FOC: Fusarium oxysporum f. sp. cubense ; VCG: Vegetative Compatibility Groups; IGC: Inter Genic Spacer; CD: Conditionally Dispensable; TE: transposable Elements; IR: Inverted Repeats; HCT: Horizontal Chromosome Transfer; TCS: Two-Component Systems; HK: Histidine kinase; with extracellular RR: Response Regulator; MAPK: Mitogen-Activated Protein Kinases
Introduction
Fusarium oxysporum f. sp. cubense is the causal agent of Panama disease Bananas and plantains are very popular staple food consumed worldwide, their production and commercialization represent important source of incomes in tropical and subtropical countries of Asia, Africa, Latin America and the Caribbean [1]. Intra and interspecific crosses between Musa acuminata and Musa balbisiana.produce commercial polyploid cultivars, which are parthenocarpy and propagated by vegetative methods. This feature reduces the genetic variation, and then, the selection of cultivars with advantageous agronomic traits such as disease resistance [2]. Fusarium oxysporum f. sp. cubense (FOC) is the causal agent of vascular wilt or Panama disease, it has been the most deleterious factor that affects the banana and plantain production [3].
Foc is an ascomycete that invades the root of the host plant, once inside, it gets incorporated into the xylem vessels and spreads through the whole plant, causing yellowing on the leaves at early stages of infection; the severity gradually increases resulting in browning and death of the leaves. Translocation of the fungus from the root causes discoloration of the rhizome and browning of the vascular bundles [4] (Figure 1). Foc can be disseminated by transferring infected rhizomes used for asexual propagation of the plant, or by the generation of asexual spores such as macroconidia, microconidia and chlamydospores, which can persist into the soil for years until a new host plants are sensed by this pathogen [5]. An outbreak in the early 90 of Foc with wider host range (Fusarium oxysporum f. sp. cubense Tropical Race 4, TR4) has been extended in Asia, Australia and Africa [6]. Nowadays, the worldwide banana production has been hampered by the Panama Disease caused by TR4, additionally, import bans imposed by phytosanitary policies affect incomes of producers in developing countries [1]. Because of the clonal nature of commercial banana cultivars, the introgression of resistance by conventional breeding does not seem to be a reliable approach to abrogate the disease. Research aimed to clarify the cause and origin of this race, and the molecular basis of its enhanced and broader virulence is necessary to generate biotechnology-based approaches of crop protection.
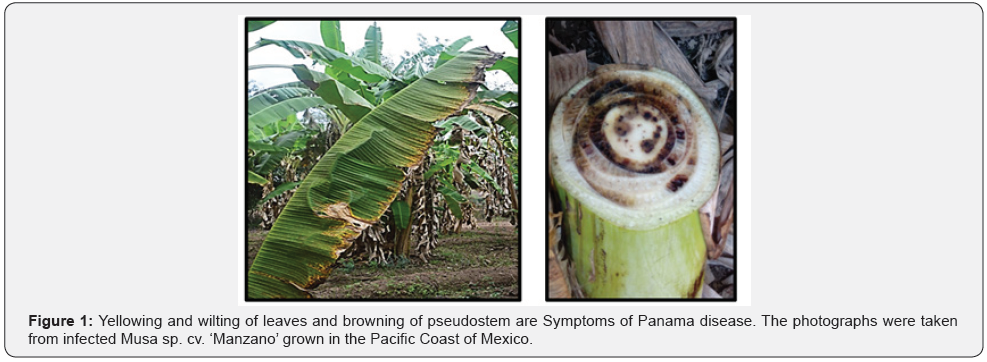
Classification and variability of Fusarium oxysporum f. sp. cubense
The Fusarium oxysporum species complex (FOSC) is a polyphyletic group without sexual stage that comprises a ubiquitous soil borne pathogens and non-pathogenic fungi. As an alternative to generate variability, filamentous fungi, including the FOSC, often exchange nuclei by anastomosis, a fusion of compatible hyphae that produces cells with 2 haploid nuclei, a condition known as heterokaryosis [7-9]. The exchange of genetic material gives hints about the evolutionary origin of specific isolates. FOSC isolates are further classified into Vegetative Compatibility Groups (VCGs), two isolates share the same group when are able to anastomose and form heterokaryons. An independent VCG encompasses a particular clonal lineage that correlates to the geographical distribution. According to this criterion, there are currently 24 VCGs of Foc [10]. Anastomosis and processes of DNA uptake are facilitated by ecological proximity between organisms and makes the horizontal gene transfer possible [11]. Therefore, horizontal gene transfer could minimize the effects of lack of sexual reproduction in members of the FOSC because allows the acquisition of novel molecular mechanisms necessary to cause disease in selected hosts and counteract the response of resistant cultivars.
Every VCG has undergone a particular evolutionary history that caused the acquisition of virulence. The term forma specialis (f. sp.) denotes the host range of a particular strain, which is normally limited to one plant species [12]. For instance, Fusarium oxysporum f. sp. lycopersici and Fusarium oxysporum f. sp. asparagi cause disease in tomato and asparagus, respectively. Thus, Foc is a specialist pathogen of Musa spp., but it is further classified into races that indicate the susceptible banana cultivars affected. Foc Race 1 (Foc1) infects cultivars of the sub group ‘Gros Michel’ derived from Musa acuminata ; and cultivars of the subgroup ’Silk’ and ‘Pome’ derived from crosses Musa acuminata X Musa balbisiana [3]. Foc1 destroyed the production of ‘Gros Michel’ bananas during the XIX century in Panama, causing severe economic losses [13], it is still a disease problem where those banana subgroups are produced. The replacement by the resistant cultivar ‘Cavendish’ was the only way to abolish Panama Disease caused by Foc1. Foc Race 2 (Foc2) infects the ‘Bluggoe’ cultivar derived from crosses Musa acuminata X Musa balbisiana.. Foc Race 3 (Foc3) infects species from genus Heliconia. Foc Race 4 (Foc4) is divided into tropical race 4 (TR4) that infects ‘Cavendish’ in tropical conditions, and Subtropical Race 4 (STR4) which can only infect ‘Cavendish’ in subtropical conditions. There are not common VCGs among Foc1 and Foc4 [3,10,14].
Then, the ability to infect resistant plants of Foc4 was a process independent of the evolutionary history of Foc1. The introduction of the Foc1-resistant cultivar ‘Cavendish’ restricted the epidemic of the Panama disease, it was not considered as a threat until early 90, when plantations in the Southeast Asia of ‘Cavendish’ started to be affected by TR4, that afterwards was dispersed in Australia, Africa and the Middle East [3]. So far, there are no reports of the presence of TR4 in Latin America and the Caribbean, but eventually, it will arrive, farmers have to prepared to face it. TR4 cause disease in the in tropical and subtropical conditions, but also displays enhanced colonization in the hosts of Foc1 [15]. Identification of TR4 isolates or any other formae especiales by morphological criteria is unviable, PCR and sequencing-based methodologies are the convincing evidences for detecting TR4. For example, the Intergenic spacer region (IGS) of ribosomal operons or the translation elongation factor 1-alpha (EF-1α) are markers recommended to differentiate Foc1 and Foc4 [16]. Knowledge of the virulence factors of distinct isolates of Foc4 will help in the design more accurate methods of diagnostics.
Comparative genomics reveals potential differences in the virulence of races of Fusarium oxysporum f. sp. cubense
The availability of genetic information of causative agents of the Panama disease allows large-scale comparisons of their genes, and therefore, identify the molecular mechanisms of TR4 that circumvent the resistance of ‘Cavendish’ or suppress a sort of plant immune responses. Comparative genomics revealed that Foc1 strain N2 and TR4 (reference strain B2) have genome sizes of 47.8Mb and 53.1Mb, respectively. Both races share 15140 orthologous genes, the difference in the genome size is reflected in the total of protein-coding genes, 17462 genes in Foc1 and 18065 genes in TR4 [17]. The higher number of unique genes in TR4 might entail a gain of function effect that allow infection in ‘Cavendish’ and also strengthen disease to the hosts of Foc1. For example, when Foc accesses into the plant, they need to produce secretory proteins such as cell wall degrading enzymes whose activity provides carbon source. Based on the in-silico detection of signal peptides, there were 1298 genes encoding secreted proteins in the genome Foc1, while 1342 of these genes were identified in TR4. Production and secretion of small proteins known as effectors is crucial to the establishment of the plant infection. Effectors are phytopathogen-specific secreted proteins that interact with specific host regulators, whether in the apoplast or inside host cell; this interaction manipulates plant immune responses and other signal transduction events that permits the pathogen’s growth [17,18].
SECRETED IN XYLEM genes and their role in Fusarium oxysporum-plant interaction
Pathogenic strains of Fusarium oxysporum produce effectors known as SIX (SECRETED IN XYLEM), they are detectable in the xylem sap of tomato plants infected with Fusarium oxysporum f. sp. lycopersici (Fol) [19]. So far, there are 14 SIX genes reported (SIX1 to SIX14). These genes encode for small secreted proteins, most of them are cysteine-rich [20]. However, the confirmation of their role in pathogenicity has been only demonstrated for SIX1 in Fol and Fusarium oxysporum f.sp. conglutinans (Focg) and SIX3, SIX4, SIX5 and SIX6 in Fol [21-24]. SIX genes are absent non-pathogens strains of Fusarium oxysporum, and every forma specialis has a particular array of SIX genes, this indicative that a selective pressure given by the host might promote the adoption of selected group of secreted protein genes that provides specialization to infect a limited host range [20]. Sequencing of SIX loci of different isolates revealed that Foc1 and Foc4 contain distinct arsenal of SIX genes (Table 1).
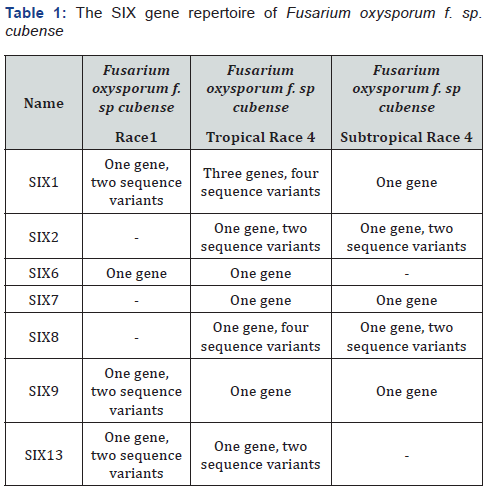
SIX genes are subject to strong selection due to polymorphisms in the corresponding interactors and by the selection of cultivars carrying potential resistance proteins, then, isolates of Foc present sequence variants in specific genes. For example, there are two sequence variants of SIX1 in isolates of Foc1, while only one sequence of SIX1 is detected in isolates of STR4 [10,17,25]. It is worth to mention that TR4 has three independent copies of SIX1 as they were identified in distinct scaffolds during the sequencing of the genome of the reference strain B2 [17]. In order to answer whether the variation found in the SIX genes of Foc is relevant, the role of such effector proteins in other pathogens has to be considered. In Fol, effector SIX1 is required during early stages of infection. Its expression is induced during root penetration and continues through invasion of xylem vessels but is not detected upon plant cell death [26]. Deletion of SIX1 in Focg (Focg-SIX1) attenuates the disease symptoms in cabbage [23]. Expression of Fol-SIX1 in Focg-SIX1 strain does not complement its reduced virulence, suggesting that substitutions in the sequence of SIX1 are needed to confer the host specificity. Interestingly, three copies of the SIX1 gene (SIX1a, SIX1b and SIX1c) are present in the genome of TR4, unlike Foc1, which has only one copy [17]. The gain of forms of SIX1 in TR4 could be a necessary adaptation to initiate the infection of the resistant ‘Cavendish’ plants. It has to be determined whether the sequence variation in the SIX1 genes of Foc1 and Foc4 implies a significant structural change in the protein that allows recognition of the host target.
SIX2 is present in STR4, TR4 and other formae speciales such as Fol, Fusarium oxysporum f. sp. niveum (pathogen of watermelon) and Fusarium oxysporum f. sp. cucumerinum (pathogen of watermelon) [20]. One particular SIX2 deletion mutant of Fol (Fol-SIX2) slightly reduced disease symptoms in tomato seedlings, but other two independent Fol-SIX2 strains did not cause disease [27]. This evidence suggests that the role of this protein is secondary, although in the case of Foc, structure and abundance of the target in banana plants has to influence the in-planta function of the SIX2 effector.
SIX8 is a multy-copy gene in Fol, and it has been detectable in formae speciales of cucurbits [20]. PCR-based analysis in different isolates illustrated that SIX8 is unique of Foc4 and the sequence variation in SIX8 can be useful in distinguishing TR4 and STR4, but it is unknown whether such variations affects structure of the effector, or whether SIX8 is required to abolish the resistance of ‘Cavendish’ plants [25]. Selected group of SIX proteins are recognized as avirulence factors in resistant cultivars. The effector SIX1 (AVR3) induced resistance in plants carrying the I-3 gene from the wild tomato Solanum pennellii [28]. SIX3 (AVR2) is introduced into the host cell and coupled with SIX5 triggers resistance and cell death in tomato plants carrying the I-2 gene [29].
This pair of effectors interact at the plasmodesmata of susceptible cultivars as a potential mechanism to distribute AVR2 in the host tissue [24]. I-2 mediated cell death upon delivery of SIX3/AVR2 is suppressed by SIX6 when co-expressed in the heterologous system Nicotiana benthamiana [21]. The genes RGC2 and RGC5 from the wild banana Musa acuminata ssp. malacciensis are candidate Foc4-resistance genes [30]. The structure of RGC2 and RGC5 proteins is reminiscent to the I-2 resistance protein that confers resistance to Fol carrying SIX3 (AVR2). Expression of RGC2 in transgenic ‘Cavendish’ banana plants confers resistance to TR4 [31].
The SIX3 and SIX5 proteins that trigger resistance in I-2 plants are lacking in Foc4 (either TR4 or STR4), in this case, another intracellular effector might elicit such race specific resistance. Comparative analysis of the genomes of different races will be useful in determine the avirulence factors of Foc4. Deletion of SIX9 did not affect the virulence of a representative strain of Fusarium oxysporum f.sp radicis-cucumerinum (Forc016) in cucurbit plants [32]. So far, there are no reports of the function of SIX7, SIX9 and SIX13 in the execution of the Panama disease. The role of these effector has to be clarified in the future.
Taken together, these results indicate that SIX proteins are functionally linked, thus, the role of SIX proteins in Foc could be considered as a whole, since the presence of two or more specific SIX genes might be a condition to establish the disease or trigger resistance. Furthermore, the selection of TR4 and STR4 could be favored by the gain of additional SIX genes whose molecular function might subvert the resistance of ‘Cavendish’ plants. Therefore, the understanding of the underlying mechanisms of acquisition of SIX and other virulence genes is necessary to decipher the biogenesis or emergence of novel causal agents of Panama disease.
SIX genes are encoded at conditionally dispensable chromosomes
Besides the core genome, the presence of supernumerary or accessory chromosomes is a common feature of pathogenic fungi. Those chromosomes are referred to as Conditionally Dispensable (CD) because they are not involved in saprophytic growth but give certain adaptive advantages in a limited number of strains [33]. High abundance of Transposable Elements (TE), either autonomous or non-autonomous, is a hallmark of CD chromosomes. TE activity causes DNA translocation, and also deletions, inversions and duplications. All these events could be involved in the construction of CD chromosomes and influence the specific combination of SIX and other virulence genes [34,35].
Mechanisms underlying the formation of CD chromosomes are independent of the origin of the core genome, however, the presence of specific virulence genes and exclusion of avirulence factors confer the ability to colonize specific hosts. 40% of the genome of the tomato pathogen Fol4287 is lineage-specific, particularly, the chromosome 14 (chr14) is a CD chromosome that harbors the SIX genes and TE [34]. The chr14 can be completely lost, or undergo deletions and duplications during vegetative growth, suggesting that DNA breaks produced by active TE encoded within CD chromosomes can also destabilize their structure, and thus, compromise the virulence, but, the saprophytic growth is not affected as no housekeeping genes are encoded into CD chromosomes, [36]. In vitro experiments revealed that the chr14 can be specifically transferred from a virulent to an avirulent strain, conferring the ability to infect tomato plants. Miniature impala (mimp) is a non-autonomous TE located at the promoters of SIX genes in Fol4287 [34,37].
The presence of mimp does not affect the expression of SIX genes, it rather locates at this position as consequence of the recombination of their Inverted Repeats (IR) with a homolog or similar sequence. When a SIX gene is encoded in the nearby of the IR, it can be mobilized during transposition together with the mimp, this event could favor the formation of a miniclusters in Fol4287, one formed by SIX1-SIX2 together with the salicylate hydrolase homolog SHH, and another containing SIX3 and SIX5 [37]. Detection of genes in the vicinity of mimp sequences is currently an approach to search for effector genes, it has led the identification of the avirulence gene AVRFOM2 in Fusarium oxysporum f. sp melonis [38]. The mimp element does not influence the expression of SIX genes, however, the sequence AAGTCGGCAGTT is frequently found in the promoter regions between the mimp and the SIX coding region and is functional for its in planta expression [37,39].
In Forc016, a CD chromosome (pathogenicity chromosome, chrRC), carries SIX genes, it is required to infect cucurbit plants [32]. The proximity between SIX genes and mimp was also detected, which confirms the correlation between both sequences. This pathogenicity chromosome can be mobilized to a non-pathogenic strain to enable the infection of cucurbit plants, which exhibits the functionality and biological impact of chrRC in a newly receptor strain. As in the case of the above mentioned Fol4287, chrRC is specifically transferred, and there are no another detectable DNA sequences from the original virulent strain. This potential Horizontal Chromosome Transfer (HCT) has to be considered as a mechanism that spreads virulence among organisms.
Sequence analysis of several markers revealed that Foc1 has a polyphyletic origin, encompassing five clonal lineages that adopted the virulence towards banana [40]. Electrophoretic karyotype demonstrated differences in the number of chromosomes and genome sizes in the VCGs that constitute Foc1 [40,41]. The number of chromosomes varies from 9 to 14, even there are alteration in the number and molecular weight of chromosomes in strains that belong to the same VCG. Such variations might be indicative of horizontal transfer of CD chromosomes and their corresponding DNA rearrangements mediated by activity of TE. Although there are not evidences of CD chromosomes in TR4, the larger genome size together with the higher number or genes might be gained by a mechanism of HCT that allowed the colonization of resistant plants. PCRbased detection of SIX genes in isolates of Foc1 and Foc4 (both STR4 and TR4) can be considered an indirect evidence of CD chromosomes [10,25,42].
Besides FOSC, another species that infect flower bulbs such as Fusarium proliferatum, Fusarium hostae and Fusarium agapanthi contain SIX genes with highly similar nucleotide identity to the homologous genes of Fusarium oxysporum. This finding, and the identification of mimp elements in the genomes of these species are evidences of inter-species horizontal transfer, possibly facilitated by the conditions of the shared habitat where the fungi interact [43]. Therefore, the molecular origin of the virulence in Fusarium oxysporum and the emergence of the formae especiales might be a consequence of the DNA shuffling that compiles SIX genes and other virulence genes into CD chromosomes, which can also be mobilized into non-pathogenic strains. Research focused on these processes in causal agents of Panama Disease will contribute to our understanding about the biogenesis of Foc1 and Foc4 as a consequence of formation and mobilization of CD chromosomes, and to decipher whether the transfer of SIX genes into another members of the same VCG or another species is a factor that generates novel races of pathogens.
Potential regulatory mechanisms of SIX gene expresin Fusarium oxysporum f. sp. cubense
In order to be functional, structure of genes acquired by HCT has to be compatible to the receptor organism. Their promoters has to be recognized by transcription factors, the introns has to be properly removed by the splicing machinery, and the codon usage of the transcripts should favor the accumulation of the protein. During the curse of the formation of CD chromosomes, specific regulators such as transcription factors might be included in conjunction with the SIX genes as an evolutionary process that ensure the expression of SIX and other effector genes. Furthermore, SIX genes and potential transcription factors acquired by HCT have to be coupled into the signal transduction pathways triggered along infection. Specific signal transduction pathways mediate the transition from saprophytic lifestyle to pathogenesis once the pathogen senses the host and accesses into the root. Perception of the host, or host-derived molecules might be mediated by sensors of extracellular stimuli such as Two-Component Systems (TCS). TCS are modules composed by a transmembrane Histidine kinase (HK) with an extracellular domain, and a response regulator (RR) with transcriptional activity [44]. The extracellular domain senses the stimulus and promotes the autophosphorylation of the HK, which transfers the phosphoryl group to the RR. There are encoded 20 genes of HK and 3 RR genes in the genomes of Foc1 and TR4. The activity of HK and RR probably converge in regulate the expression of SIX genes. In the wheat pathogen Fusarium graminearum, and other fungi, TCS and Mitogen-Activated Protein Kinases (MAPK) are linked [45]. MAPK are sequentially activated protein kinases that typically phosphorylate transcriptional regulators. In FolATCC66044, silencing of MAPK gene FMK1 reduced the virulence in tomato [46]. Targets of FMK1 has to be identified, however, if the virulence is reduced in silenced strain, the activity of FMK1 might be directed to regulate the genes of the CD chromosomes, including SIX genes, but intermediate step involving transcription factors have to be considered as part of this regulatory process.
Besides the perception of the host plant, Transcription Factors (TF) are required to mediate the transcriptional reprograming to establish the pathogenic growth and cause symptoms. Activity of such transcription factors, either encoded into the core genome or into CD chromosomes, have to correlate to the composition of cis-acting elements into the promoters of SIX genes and other virulence factors. In Fol4287, the transcription factor SIX GENE EXPRESSION 1 [SGE1] has a WOPR DNA-binding domain, it is involved in regulating the expression of SIX1, SIX2, SIX3 and SIX5 genes [47]. In accordance to its transcriptional function, the deletion of SGE1 causes loss of pathogenicity on tomato plants. SGE1 is located at core genome, and the conservation of this gene in other fungi suggests that SGE1 is a conserved transcription factor that evolved as a regulator of SIX genes gained by horizontal transfer. Recently, the homolog of SGE1 has been identified in TR4, it is involved colonization of banana roots and pathogenicity [47,48]. The role of SGE1 in the transcription of SIX genes in Foc1 and Foc4 has to be confirmed in the future as a way to demonstrate that SGE1-mediated expression of SIX genes is a determining factor that enables virulence.
The Fusarium Transcription Factor (FTF) belong to the Zn2Cys6 family of transcription factors. The knockdown of FTF1and FTF2 reduced virulence of Fusarium oxysporum f. sp. phaseoli (Fop) towards bean plants [49]. Down-regulation of FTF1/2 in Fop also decreases the in-planta expression of SIX1, SIX6 and the homolog of SGE1, but their role as direct regulators of SGE1 and SIX genes has to be defined. Selected FTF genes such as FTF1, are encoded into the chr14 of Fol4287, silencing of FTF1/2 genes also attenuates expression of SIX1 and SIX6 as well as disease progress [49]. The FTF1 gene of Fol4187 and have a mimp at the promoter region, which resembles the structure of SIX genes [37,49]. The sequence T(G/A)CCG is the consensus binding site of FTF1 [37,39] which overlaps with the sequence element at the promoters of SIX genes identified by Schmidt and co-workers . In the case of Fol4287, both FTF1 and SGE1 are required to induce expression of SIX1 and SIX13, revealing that the expression of SIX genes requires cooperative activity of transcription factors from the core genome and the CD chromosome [39].
In order to start with the characterization of transcription factors in the expression of SIX genes of Foc, it is worth to mention that the genome of Foc1 and TR4 harbors 346 and 355 Zn2Cys6 genes, respectively [17]. This number of genes is markedly higher compared to Neurospora crassa, which only carries 45 genes of this family [50]. The expansion of Zn2Cys6 genes in Foc might be an adaptation that facilitated the expression of SIX and other effector genes. The identification of the subset of Zn2Cys6 genes carried on the accessory CD chromosomes and on the core genome of Foc, as well as the characterization of their function is necessary to understand the similarities and differences on the regulatory mechanisms of SIX gene expression and to clarify whether a mechanism mediated by transcription factors is critical to abrogate the resistance of ‘Cavendish’ plants.
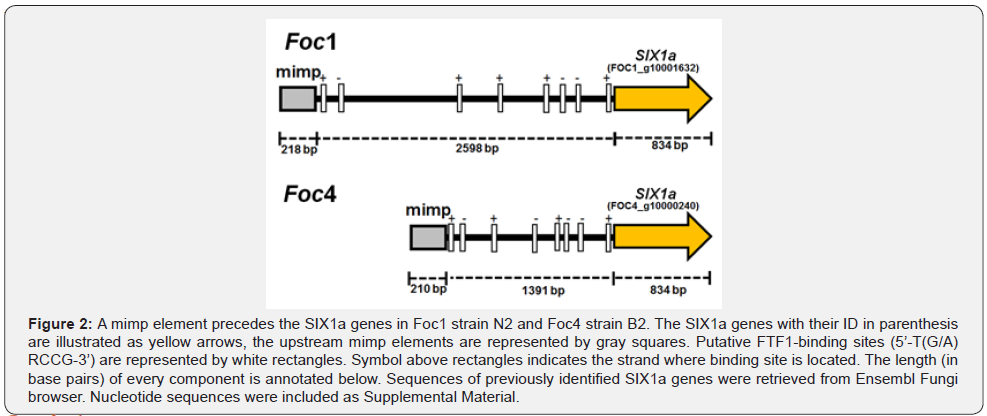
Genomic context of Foc1-SIX1a and Foc4-SIX1a is reminiscent to the SIX genes encoded into CD chromosomes
Incongruence between the phylogenies of SIX genes and marker genes used for phylogenetic reconstruction such as TEF- 1α suggests the SIX genes were horizontally inherited in Foc [10]. The context of Six1a gives further evidences about the potential horizontal transfer of SIX genes in races of Foc. SIX1a is widely distributed in isolates of Foc1 and Foc4 and represents a reliable model to investigate the horizontal transmission of virulence genes [10, 51]. Likewise, the SIX genes of Fol4287 and Forc016 located at CD chromosome, the SIX1a of reference strains is accompanied by a upstream mimp element (Figure 2). Since FTF1 is required to induce the expression of SIX genes in Fol4287 [39], putative FTF1-binding sites were searched for at the DNA sequence surrounded by the mimp and the start codon of SIX1a. Several putative FTF1 binding sites were identified both in (+) and (-) DNA strands, suggesting that in planta expression SIX1a is potentially regulated by the homolog of FTF1. Although SGE1 influences expression of SIX genes in Fol4287, and its role in the pathogenicity of Foc4 is reported [39,48], SGE1-binding sites were not identified at the promoters of Foc1-SIX1a and Foc4- SIX1a. These findings illustrate that SIX genes share a genetic structure independent of the forma specialis that might allow the recognition of FTF1 and other Zn2Cys6 factors at different position of those promoters to regulate transcription during the infection. Nucleotide sequences of SIX1a genes, promoters and mimp elements were included as Supplemental Material.
Conclusion
Nowadays, the Panama disease, specially caused by the TR4, is the main factor that hampers the worldwide production of bananas. Fusarium oxysporum is prone to acquire DNA by horizontal gene transfer as a mechanism to compensate the lack of sexual reproduction. Transfer of complete CD chromosomes carrying TE, SIX virulence genes and their corresponding transcriptional regulators has been reported in selected formae especiales of FOSC, such process is the driving force of the evolution of pathogenic strains because it confers the ability to infect a narrow host range. This mechanism likely entailed the selection of Foc races as banana pathogens, including TR4, a race with improved repertoire SIX and other effector genes that might enhance the virulence and allow the infection of resistant plants. The analysis of complete genome sequences of isolates of Foc coming from different geographic locations will be helpful to understand how Foc is reinventing itself by forming and mobilizing CD chromosomes carrying specific combination of sequence variants of SIX genes.
Acknowledgement
LDMB thanks to the “Cátedras CONACYT Program” (Grant 538) for the financial support. “Programa PRODEP UMAR CA- 19” supported JLVO, and “Programa PRODEP UMAR CA-24” supported MACO and ACSE.
To Know More
About Journal of
Medical Microbiology and Diagnosis-Biotechnology Open Access Journals Please Click on: https://juniperpublishers.com/aibm/index.php
To Know More About Open
Access Journals Publishers Please Click on: Juniper
Publishers



No comments:
Post a Comment